Prediction of Second-Order Rate Constants of Sulfate Radical with Aromatic Contaminants Using Quantitative Structure-Activity Relationship Model
Abstract
:1. Introduction
2. Materials and Methods
2.1. Dataset
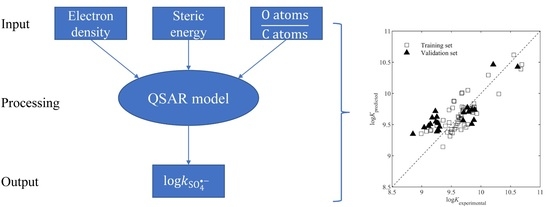
2.2. QSAR Model Development and Characterization
2.3. Validation of the Model
2.4. Relative Contribution of Each Descriptor
2.5. Applicability Domain
3. Results
3.1. Selection of Significant Descriptors
3.2. Exclusion of Outliers
3.3. Statistical Characteristics of the Developed Model
4. Discussion
4.1. Interpretation of the Model
4.2. Applicability Domain
4.3. Comparison with Other Models
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xiao, R.; Luo, Z.; Wei, Z.; Luo, S.; Spinney, R.; Yang, W.; Dionysiou, D.D. Activation of peroxymonosulfate/persulfate by nanomaterials for sulfate radical-based advanced oxidation technologies. Curr. Opin. Chem. Eng. 2018, 19, 51–58. [Google Scholar] [CrossRef]
- Oh, W.-D.; Dong, Z.; Lim, T.-T. Generation of sulfate radical through heterogeneous catalysis for organic contaminants removal: Current development, challenges and prospects. Appl. Catal. B-Environ. 2016, 194, 169–201. [Google Scholar] [CrossRef]
- Zhou, D.; Zhang, H.; Chen, L. Sulfur-replaced Fenton systems: Can sulfate radical substitute hydroxyl radical for advanced oxidation technologies? J. Chem. Technol. Biotechnol. 2015, 90, 775–779. [Google Scholar] [CrossRef]
- Oh, W.-D.; Lua, S.-K.; Dong, Z.; Lim, T.-T. Performance of magnetic activated carbon composite as peroxymonosulfate activator and regenerable adsorbent via sulfate radical-mediated oxidation processes. J. Hazard. Mater. 2015, 284, 1–9. [Google Scholar] [CrossRef]
- Wacławek, S.; Lutze, H.V.; Grübel, K.; Padil, V.V.T.; Černík, M.; Dionysiou, D.D. Chemistry of persulfates in water and wastewater treatment: A review. Chem. Eng. J. 2017, 330, 44–62. [Google Scholar] [CrossRef]
- Su, H.; Yu, C.; Zhou, Y.; Gong, L.; Li, Q.; Alvarez, P.J.J.; Long, M. Quantitative structure-activity relationship for the oxidation of aromatic organic contaminants in water by TAML/H2O2. Water Res. 2018, 140, 354–363. [Google Scholar] [CrossRef]
- Seo, J.-S.; Keum, Y.-S.; Li, Q.X. Bacterial degradation of aromatic compounds. Int. J. Env. Res. Public Health 2009, 6, 278–309. [Google Scholar] [CrossRef]
- Fuchs, G.; Boll, M.; Heider, J. Microbial degradation of aromatic compounds—From one strategy to four. Nat. Rev. Microbiol. 2011, 9, 803–816. [Google Scholar] [CrossRef]
- Pari, S.; Wang, I.A.; Liu, H.; Wong, B.M. Sulfate radical oxidation of aromatic contaminants: A detailed assessment of density functional theory and high-level quantum chemical methods. Environ. Sci.-Process Impacts 2017, 19, 395–404. [Google Scholar] [CrossRef] [Green Version]
- Neta, P.; Huie, R.E.; Ross, A.B. Rate Constants for Reactions of Inorganic Radicals in Aqueous Solution. J. Phys. Chem. Ref. Data 1988, 17, 1027–1284. [Google Scholar] [CrossRef]
- Ding, H.; Hu, J. Degradation of ibuprofen by UVA-LED/TiO2/persulfate process: Kinetics, mechanism, water matrix effects, intermediates and energy consumption. Chem. Eng. J. 2020, 397, 125462. [Google Scholar] [CrossRef]
- Ye, T.; Wei, Z.; Spinney, R.; Tang, C.-J.; Luo, S.; Xiao, R.; Dionysiou, D.D. Chemical structure-based predictive model for the oxidation of trace organic contaminants by sulfate radical. Water Res. 2017, 116, 106–115. [Google Scholar] [CrossRef] [PubMed]
- Dudek, A.Z.; Arodz, T.; Galvez, J. Computational Methods in Developing Quantitative Structure-Activity Relationships (QSAR): A Review. Comb. Chem. High Throughput Screen 2006, 9, 213–228. [Google Scholar] [CrossRef]
- Liu, W.; Wang, X.; Zhou, X.; Duan, H.; Zhao, P.; Liu, W. Quantitative structure-activity relationship between the toxicity of amine surfactant and its molecular structure. Sci. Total Environ. 2020, 702, 134593. [Google Scholar] [CrossRef] [PubMed]
- Nolte, T.M.; Chen, G.; van Schayk, C.S.; Pinto-Gil, K.; Hendriks, A.J.; Peijnenburg, W.J.G.M.; Ragas, A.M.J. Disentanglement of the chemical, physical, and biological processes aids the development of quantitative structure-biodegradation relationships for aerobic wastewater treatment. Sci. Total Environ. 2020, 708, 133863. [Google Scholar] [CrossRef]
- Selassie, C.; Verma, R.P. History of quantitative structure-activity relationships. Burg. Med. Chem. Drug Discov. 2003, 1, 1–96. [Google Scholar]
- Li, C.; Zheng, S.; Li, T.; Chen, J.; Zhou, J.; Su, L.; Zhang, Y.-N.; Crittenden, J.C.; Zhu, S.; Zhao, Y. Quantitative structure-activity relationship models for predicting reaction rate constants of organic contaminants with hydrated electrons and their mechanistic pathways. Water Res. 2019, 151, 468–477. [Google Scholar] [CrossRef]
- Huang, Y.; Li, T.; Zheng, S.; Fan, L.; Su, L.; Zhao, Y.; Xie, H.-B.; Li, C. QSAR modeling for the ozonation of diverse organic compounds in water. Sci. Total Environ. 2020, 715, 136816. [Google Scholar] [CrossRef]
- Li, T.; Huang, Y.; Wei, G.; Zhang, Y.-N.; Zhao, Y.; Crittenden, J.C.; Li, C. Quantitative structure-activity relationship models for predicting singlet oxygen reaction rate constants of dissociating organic compounds. Sci. Total Environ. 2020, 735, 139498. [Google Scholar] [CrossRef]
- Borhani, T.N.G.; Saniedanesh, M.; Bagheri, M.; Lim, J.S. QSPR prediction of the hydroxyl radical rate constant of water contaminants. Water Res. 2016, 98, 344–353. [Google Scholar] [CrossRef] [PubMed]
- Xiao, R.; Ye, T.; Wei, Z.; Luo, S.; Yang, Z.; Spinney, R. Quantitative Structure-Activity Relationship (QSAR) for the Oxidation of Trace Organic Contaminants by Sulfate Radical. Environ. Sci. Technol. 2015, 49, 13394–13402. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, D.M. The Problem of Overfitting. J. Chem. Inf. Comput. Sci. 2004, 44, 1–12. [Google Scholar] [CrossRef] [PubMed]
- OECD. Guidance Document on the Validation of (Quantitative) Structure-Activity Relationship [(Q) SAR] Models; Organisation for Economic Co-Operation and Development: Paris, France, 2007. [Google Scholar]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16 Rev. A.03; Gausian: Wallingford, CT, USA, 2016. [Google Scholar]
- DeTar, D.F. Calculation of formal steric enthalpy with MM2. J. Org. Chem. 1992, 57, 902–910. [Google Scholar] [CrossRef]
- Craney, T.A.; Surles, J.G. Model-Dependent Variance Inflation Factor Cutoff Values. Qual. Eng. 2002, 14, 391–403. [Google Scholar] [CrossRef]
- Stine, R.A. Graphical Interpretation of Variance Inflation Factors. Am. Stat. 1995, 49, 53–56. [Google Scholar]
- Sudhakaran, S.; Calvin, J.; Amy, G.L. QSAR models for the removal of organic micropollutants in four different river water matrices. Chemosphere 2012, 87, 144–150. [Google Scholar] [CrossRef]
- Gramatica, P. Principles of QSAR models validation: Internal and external. QSAR Comb. Sci. 2007, 26, 694–701. [Google Scholar] [CrossRef]
- Tropsha, A.; Gramatica, P.; Gombar, V.K. The Importance of Being Earnest: Validation is the Absolute Essential for Successful Application and Interpretation of QSPR Models. QSAR Comb. Sci. 2003, 22, 69–77. [Google Scholar] [CrossRef]
- Golbraikh, A.; Tropsha, A. Beware of q2! J. Mol. Graph. Model. 2002, 20, 269–276. [Google Scholar] [CrossRef]
- Kaneko, H. Estimation of predictive performance for test data in applicability domains using y-randomization. J. Chemom. 2019, 33, e3171. [Google Scholar] [CrossRef]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Netzeva, T.I.; Worth, A.P.; Aldenberg, T.; Benigni, R.; Cronin, M.T.D.; Gramatica, P.; Jaworska, J.S.; Kahn, S.; Klopman, G.; Marchant, C.A.; et al. Current Status of Methods for Defining the Applicability Domain of (Quantitative) Structure-Activity Relationships: The Report and Recommendations of ECVAM Workshop 521,2. ATLA-Altern. Lab. Anim. 2005, 33, 155–173. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, A.C. Plots, Transformations and Regression: An Introduction to Graphical Methods of Diagnostic Regression Analysis. 1985. Available online: https://books.google.rs/books/about/Plots_Transformations_and_Regression.html?id=L1SqAAAAIAAJ&redir_esc=y (accessed on 25 June 2021).
- Liu, Y.; Cheng, Z.; Liu, S.; Tan, Y.; Yuan, T.; Yu, X.; Shen, Z. Quantitative structure activity relationship (QSAR) modelling of the degradability rate constant of volatile organic compounds (VOCs) by OH radicals in atmosphere. Sci. Total Environ. 2020, 729, 138871. [Google Scholar] [CrossRef]
- Montgomery, D.; Runger, G. Applied Statistics and Probability for Engineers; John Wily and Sons, Inc.: Hoboken, NJ, USA, 2018. [Google Scholar]
- Ding, H.; Chen, C.; Zhang, X. Linear solvation energy relationship for the adsorption of synthetic organic compounds on single-walled carbon nanotubes in water. SAR QSAR Environ. Res. 2016, 27, 31–45. [Google Scholar] [CrossRef]
- Ashby, E.C. Single-electron transfer, a major reaction pathway in organic chemistry. An answer to recent criticisms. Acc. Chem. Res. 1988, 21, 414–421. [Google Scholar] [CrossRef]
- Luo, S.; Wei, Z.; Dionysiou, D.D.; Spinney, R.; Hu, W.-P.; Chai, L.; Yang, Z.; Ye, T.; Xiao, R. Mechanistic insight into reactivity of sulfate radical with aromatic contaminants through single-electron transfer pathway. Chem. Eng. J. 2017, 327, 1056–1065. [Google Scholar] [CrossRef]
- Siafarika, P.; Kouderis, C.; Kalampounias, A.G. Non-Debye segmental relaxation of poly-N-vinyl-carbazole in dilute solution. Mol. Phys. 2021, 119, e1802075. [Google Scholar] [CrossRef]
- Jaworska, J.; Nikolova-Jeliazkova, N.; Aldenberg, T. QSAR Applicability Domain Estimation by Projection of the Training Set in Descriptor Space: A Review. ATLA-Altern. Lab. Anim. 2005, 33, 445–459. [Google Scholar] [CrossRef]
- O’Neill, P.; Steenken, S.; Schulte-Frohlinde, D. Formation of radical cations of methoxylated benzenes by reaction with hydroxyl radicals, thallium(2+), silver(2+), and peroxysulfate (SO4.−) in aqueous solution. Optical and conductometric pulse radiolysis and in situ radiolysis electron spin resonance study. J. Phys. Chem. 1975, 79, 2773–2779. [Google Scholar]
- Steenken, S.; O’Neill, P.; Schulte-Frohlinde, D. Formation of radical zwitterions from methoxylated benzoic acids. 1. One electron oxidation by thallium(2+), silver(2+), and sulfate(1−) ions. J. Phys. Chem. 1977, 81, 26–30. [Google Scholar] [CrossRef]
- Choure, S.C.; Bamatraf, M.M.M.; Rao, B.S.M.; Das, R.; Mohan, H.; Mittal, J.P. Hydroxylation of Chlorotoluenes and Cresols: A Pulse Radiolysis, Laser Flash Photolysis, and Product Analysis Study. J. Phys. Chem. A 1997, 101, 9837–9845. [Google Scholar] [CrossRef]
- Zhang, T.; Chen, Y.; Leiknes, T. Oxidation of Refractory Benzothiazoles with PMS/CuFe2O4: Kinetics and Transformation Intermediates. Environ. Sci. Technol. 2016, 50, 5864–5873. [Google Scholar] [CrossRef] [Green Version]
- Canle López, M.; Fernández, M.I.; Rodríguez, S.; Santaballa, J.A.; Steenken, S.; Vulliet, E. Mechanisms of Direct and TiO2-Photocatalysed UV Degradation of Phenylurea Herbicides. Chemphyschem 2005, 6, 2064–2074. [Google Scholar] [CrossRef]
- Kilic, M.Y.; Abdelraheem, W.H.; He, X.; Kestioglu, K.; Dionysiou, D.D. Photochemical treatment of tyrosol, a model phenolic compound present in olive mill wastewater, by hydroxyl and sulfate radical-based advanced oxidation processes (AOPs). J. Hazard. Mater. 2019, 367, 734–742. [Google Scholar] [CrossRef] [PubMed]
- Neta, P.; Madhavan, V.; Zemel, H.; Fessenden, R.W. Rate constants and mechanism of reaction of sulfate radical anion with aromatic compounds. J. Am. Chem. Soc. 1977, 99, 163–164. [Google Scholar] [CrossRef]
- Kwon, M.; Kim, S.; Yoon, Y.; Jung, Y.; Hwang, T.-M.; Lee, J.; Kang, J.-W. Comparative evaluation of ibuprofen removal by UV/H2O2 and UV/S2O82− processes for wastewater treatment. Chem. Eng. J. 2015, 269, 379–390. [Google Scholar] [CrossRef]
- Li, A.; Wu, Z.; Wang, T.; Hou, S.; Huang, B.; Kong, X.; Li, X.; Guan, Y.; Qiu, R.; Fang, J. Kinetics and mechanisms of the degradation of PPCPs by zero-valent iron (Fe°) activated peroxydisulfate (PDS) system in groundwater. J. Hazard. Mater. 2018, 357, 207–216. [Google Scholar] [CrossRef]
- Real, F.J.; Acero, J.L.; Benitez, J.F.; Roldan, G.; Casas, F. Oxidation of the emerging contaminants amitriptyline hydrochloride, methyl salicylate and 2-phenoxyethanol by persulfate activated by UV irradiation. J. Chem. Technol. Biotechnol. 2016, 91, 1004–1011. [Google Scholar] [CrossRef]
- Rickman, K.A.; Mezyk, S.P. Kinetics and mechanisms of sulfate radical oxidation of β-lactam antibiotics in water. Chemosphere 2010, 81, 359–365. [Google Scholar] [CrossRef]
- Toth, J.E.; Rickman, K.A.; Venter, A.R.; Kiddle, J.J.; Mezyk, S.P. Reaction Kinetics and Efficiencies for the Hydroxyl and Sulfate Radical Based Oxidation of Artificial Sweeteners in Water. J. Phys. Chem. A 2012, 116, 9819–9824. [Google Scholar] [CrossRef]
- Sharma, S.B.; Mudaliar, M.; Rao, B.S.M.; Mohan, H.; Mittal, J.P. Radiation Chemical Oxidation of Benzaldehyde, Acetophenone, and Benzophenone. J. Phys. Chem. A 1997, 101, 8402–8408. [Google Scholar] [CrossRef]
- Mahdi Ahmed, M.; Barbati, S.; Doumenq, P.; Chiron, S. Sulfate radical anion oxidation of diclofenac and sulfamethoxazole for water decontamination. Chem. Eng. J. 2012, 197, 440–447. [Google Scholar] [CrossRef]
- Mohan, H.; Mittal, J.P. Direct evidence for H+-catalysed dehydration of fluorohydroxycyclohexadienyl radical: A pulse radiolysis study. J. Chem. Soc.-Faraday Trans. 1995, 91, 2121–2126. [Google Scholar] [CrossRef]
- Caregnato, P.; David Gara, P.M.; Bosio, G.N.; Gonzalez, M.C.; Russo, N.; Michelini, M.d.C.; Mártire, D.O. Theoretical and Experimental Investigation on the Oxidation of Gallic Acid by Sulfate Radical Anions. J. Phys. Chem. A 2008, 112, 1188–1194. [Google Scholar] [CrossRef]
- Li, H.; Liu, Y.; Tang, R.; Zhang, P.; Fu, H.; Yao, S.; Wang, W. Pulse radiolysis study on gatifloxacin—A fluoroquinolone antibiotic. Sci. China-Chem. 2012, 55, 1358–1363. [Google Scholar] [CrossRef]
- Paul, J.; Jensen, S.; Dukart, A.; Cornelissen, G. Optimization of a preparative multimodal ion exchange step for purification of a potential malaria vaccine. J. Chromatogr. A 2014, 1366, 38–44. [Google Scholar] [CrossRef]
- Lanzafame, G.M.; Sarakha, M.; Fabbri, D.; Vione, D. Degradation of Methyl 2-Aminobenzoate (Methyl Anthranilate) by H2O2/UV: Effect of Inorganic Anions and Derived Radicals. Molecules 2017, 22, 619. [Google Scholar] [CrossRef] [Green Version]
- Lian, L.; Yao, B.; Hou, S.; Fang, J.; Yan, S.; Song, W. Kinetic Study of Hydroxyl and Sulfate Radical-Mediated Oxidation of Pharmaceuticals in Wastewater Effluents. Environ. Sci. Technol. 2017, 51, 2954–2962. [Google Scholar] [CrossRef]
- Geeta, S.; Sharma, S.B.; Rao, B.S.M.; Mohan, H.; Dhanya, S.; Mittal, J.P. Study of kinetics and absorption spectra of OH adducts of hydroxy derivatives of benzaldehyde and acetophenone. J. Photochem. Photobiol. A Chem. 2001, 140, 99–107. [Google Scholar] [CrossRef]
- Acero, J.L.; Benítez, F.J.; Real, F.J.; Rodríguez, E. Degradation of selected emerging contaminants by UV-activated persulfate: Kinetics and influence of matrix constituents. Sep. Purif. Technol. 2018, 201, 41–50. [Google Scholar] [CrossRef]
- Roder, M.; Földiák, G.; Wojnárovits, L. Electron transfer from cresols to N3, BrO2, ClO2, NO2 and SO−4 radicals: Correlation between rate constants and one-electron reduction potentials. Radiat. Phys. Chem. 1999, 55, 515–519. [Google Scholar] [CrossRef]
- Tao, Y.; Monfort, O.; Brigante, M.; Zhang, H.; Mailhot, G. Phenanthrene decomposition in soil washing effluents using UVB activation of hydrogen peroxide and peroxydisulfate. Chemosphere 2021, 263, 127996. [Google Scholar] [CrossRef] [PubMed]
- Herrmann, H.; Exner, M.; Jacobi, H.W.; Raabe, G.; Reese, A.; Zellner, R. Laboratory studies of atmospheric aqueous-phase free-radical chemistry: Kinetic and spectroscopic studies of reactions of NO3 and SO4− radicals with aromatic compounds. Faraday Discuss. 1995, 100, 129–153. [Google Scholar] [CrossRef]
- Sabharwal, S.; Kishore, K.; Moorthy, P.N. Pulse radiolysis study of oxidation reactions of sulphacetamide in aqueous solutions. Radiat. Phys. Chem. 1994, 44, 499–506. [Google Scholar] [CrossRef]
- Zhang, R.; Yang, Y.; Huang, C.-H.; Zhao, L.; Sun, P. Kinetics and modeling of sulfonamide antibiotic degradation in wastewater and human urine by UV/H2O2 and UV/PDS. Water Res. 2016, 103, 283–292. [Google Scholar] [CrossRef]
- Zhou, L.; Ferronato, C.; Chovelon, J.-M.; Sleiman, M.; Richard, C. Investigations of diatrizoate degradation by photo-activated persulfate. Chem. Eng. J. 2017, 311, 28–36. [Google Scholar] [CrossRef]
- Merga, G.; Aravindakumar, C.T.; Rao, B.S.M.; Mohan, H.; Mittal, J.P. Pulse radiolysis study of the reactions of SO with some substituted benzenes in aqueous solution. J. Chem. Soc.-Faraday Trans. 1994, 90, 597–604. [Google Scholar] [CrossRef]
- Zhang, R.; Sun, P.; Boyer, T.H.; Zhao, L.; Huang, C.-H. Degradation of Pharmaceuticals and Metabolite in Synthetic Human Urine by UV, UV/H2O2, and UV/PDS. Environ. Sci. Technol. 2015, 49, 3056–3066. [Google Scholar] [CrossRef]
- Buxton, G.V.; Salmon, G.A.; Williams, J.E. The Reactivity of Biogenic Monoterpenes towards OH· and SO4− Radicals in De-Oxygenated Acidic Solution. J. Atmos. Chem. 2000, 36, 111–134. [Google Scholar] [CrossRef]
- Matta, R.; Tlili, S.; Chiron, S.; Barbati, S. Removal of carbamazepine from urban wastewater by sulfate radical oxidation. Environ. Chem. Lett. 2011, 9, 347–353. [Google Scholar] [CrossRef]
- Lu, X.; Shao, Y.; Gao, N.; Chen, J.; Deng, H.; Chu, W.; An, N.; Peng, F. Investigation of clofibric acid removal by UV/persulfate and UV/chlorine processes: Kinetics and formation of disinfection byproducts during subsequent chlor(am)ination. Chem. Eng. J. 2018, 331, 364–371. [Google Scholar] [CrossRef]






| Steps | Descriptors | Coefficient | t-Statistic | p-Value | RMSE | Decision | ||
|---|---|---|---|---|---|---|---|---|
| 1 | E | 1.5755 | 5.7113 | 0.0000 | 0.685 | 0.659 | 0.219 | Exclude logP |
| S | 0.0042 | 6.5814 | 0.0000 | |||||
| O/C | −0.1924 | −0.5424 | 0.5895 | |||||
| LogV | −0.2572 | −1.2209 | 0.2269 | |||||
| LogP | 0.0374 | 1.4067 | 0.1647 | |||||
| Constant | 1.8972 | |||||||
| 2 | E | 1.6799 | 6.2729 | 0.0000 | 0.675 | 0.653 | 0.221 | Exclude logV |
| S | 0.0039 | 6.4515 | 0.0000 | |||||
| O/C | −0.5938 | −2.7933 | 0.0070 | |||||
| LogV | −0.1412 | −0.7225 | 0.4727 | |||||
| Constant | 1.2381 | |||||||
| 3 | E | 1.6882 | 6.3344 | 0.0000 | 0.672 | 0.656 | 0.220 | Accept E, S and O/C as the significant descriptors |
| S | 0.0037 | 6.7496 | 0.0000 | |||||
| O/C | −0.6035 | −2.8559 | 0.0058 | |||||
| Constant | 0.8868 | |||||||
| Model | Goodness-of-Fit | Robustness | Predictive Ability | k | k′ | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RMSE | RMSEext | ||||||||||||
| Including outliers | 0.672 | 0.656 | 0.220 | 0.617 | 0.605 | 0.282 | 0.632 | 0.066 | 0.610 | 1.010 | 0.990 | 0.896 | 0.035 |
| Excluding outliers | 0.748 | 0.735 | 0.193 | 0.694 | 0.603 | 0.289 | 0.648 | 0.110 | 0.624 | 1.013 | 0.987 | 0.830 | 0.037 |
| Reference | Model Type | n* | Molecular Descriptors | Goodness-of-Fit | Robustness | Predictive Ability | AD | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| RMSE | RMSEext | ||||||||||
| Xiao et al. (2015) | MLR | 65 | Ratio of oxygen atoms to carbon; ELUMO and EHOMO energy gap | 0.866 | - | - | 0.86 | 0.89 | - | 0.89 | All but one compound from the validation set was outside the AD |
| Ye et al. (2017) | MLR and ANN | 75 | 32 molecular fragment descriptors | 0.88 (MLR); 0.99 (ANN) | - | - | - | - | - | 0.62 (MLR); 0.42 (ANN) | - |
| This study | MLR | 61 | Electron density, steric energy, and ratio of oxygen atoms to carbon | 0.748 | 0.735 | 0.193 | 0.694 | 0.603 | 0.289 | 0.648 | All data points of the validation set fell within the AD |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, H.; Hu, J. Prediction of Second-Order Rate Constants of Sulfate Radical with Aromatic Contaminants Using Quantitative Structure-Activity Relationship Model. Water 2022, 14, 766. https://doi.org/10.3390/w14050766
Ding H, Hu J. Prediction of Second-Order Rate Constants of Sulfate Radical with Aromatic Contaminants Using Quantitative Structure-Activity Relationship Model. Water. 2022; 14(5):766. https://doi.org/10.3390/w14050766
Chicago/Turabian StyleDing, Han, and Jiangyong Hu. 2022. "Prediction of Second-Order Rate Constants of Sulfate Radical with Aromatic Contaminants Using Quantitative Structure-Activity Relationship Model" Water 14, no. 5: 766. https://doi.org/10.3390/w14050766