Abstract
Recent advances in viral vector engineering, as well as an increased understanding of the cellular and molecular mechanism of retinal diseases, have led to the development of novel gene therapy approaches. Furthermore, ease of accessibility and ocular immune privilege makes the retina an ideal target for gene therapies. In this study, the nuclear hormone receptor gene Nr2e3 was evaluated for efficacy as broad-spectrum therapy to attenuate early to intermediate stages of retinal degeneration in five unique mouse models of retinitis pigmentosa (RP). RP is a group of heterogenic inherited retinal diseases associated with over 150 gene mutations, affecting over 1.5 million individuals worldwide. RP varies in age of onset, severity, and rate of progression. In addition, ~40% of RP patients cannot be genetically diagnosed, confounding the ability to develop personalized RP therapies. Remarkably, Nr2e3 administered therapy resulted in reduced retinal degeneration as observed by increase in photoreceptor cells, improved electroretinogram, and a dramatic molecular reset of key transcription factors and associated gene networks. These therapeutic effects improved retinal homeostasis in diseased tissue. Results of this study provide evidence that Nr2e3 can serve as a broad-spectrum therapy to treat multiple forms of RP.
Similar content being viewed by others
Introduction
Recent studies have demonstrated the potential of gene therapy to attenuate or slow the progression of previously untreatable inherited diseases [1]. Gene therapy has evolved from concept to clinic for various genetic disorders including hematological [2], immunological [3], ocular [4, 5], neurodegenerative [6], and metabolic disorders [7]. Due to its ease of accessibility and immune privilege, the eye holds potential as an ideal organ for gene therapy. The most widely accepted success of adeno-associated virus (AAV) vector-mediated ocular gene replacement is for treatment of Leber’s congenital amaurosis 2 (LCA2), a rare retinal disease due to mutations in the RPE65 gene [8, 9]. Currently, there are several ongoing clinical gene augmentation trials for other rare inherited retinal diseases [10,11,12,13]. However, several factors such as gene size, gene function, and the large number (~40%) of retinitis pigmentosa (RP) patients that cannot be genetically diagnosed present challenges for developing individual gene replacement/augmentation-based therapies. Thus, new therapeutic approaches are needed to circumvent these limitations. This study evaluates a unique approach using the nuclear hormone receptor (NHR) gene Nr2e3 as a genetic modifier and therapeutic agent to treat multiple retinal degenerative diseases. Results of this study demonstrate the power of a single genetic modifier in treating retinal diseases.
RP represents a group of inherited diseases, affecting an estimated 1 in 4000 individuals, that cause degeneration of rod and cone photoreceptor cells, leading to the severe vision loss [14, 15]. RP can be inherited through multiple modes of inheritance such as autosomal dominant (30–40% of cases), autosomal recessive (50–60% of cases), or X-linked (5–15% of cases) manner in syndromic or nonsyndromic forms [16,17,18]. Over 150 unique gene mutations have been associated with RP, making it highly heterogenic, with high variability in disease onset, severity, and progression [19,20,21,22,23].
Genetic modifiers are defined as allelic variants found within the normal population [24, 25]. Modifier genes can significantly affect disease outcomes, impacting onset, rate of progression, and severity [24, 26, 27]. Genetic modifier genes are powerful modulators that can enhance or suppress disease phenotypes [24, 27,28,29]. The direct impact of genetic modifiers has been studied extensively in several diseases including cystic fibrosis, epileptic encephalopathy, spinocerebellar ataxia type 1, spinal muscular atrophy, dystonia, and retinal degeneration where drastically altered phenotypes occur when genetic background is shifted [30,31,32,33,34,35,36,37,38]. Haider et al. discovered that shifting the rd7 mutation, a recessive mutation in NHR 2 family e, member 3, Nr2e3 that results in slow progressive retinal degeneration, onto three different genetic backgrounds resulted in complete suppression of the rd7 phenotype in all strains evaluated, and genetic mapping revealed that several modifier genes could independently account for this suppression [39]. The NHR 1 family d, member 1 (Nr1d1), a NHR gene, and cofactor of Nr2e3, was identified as one of the genetic modifiers that can ameliorate Nr2e3 associated retinal degeneration [40].
Mutations in human NR2E3 are associated with several forms of retinal degeneration that vary in phenotype and were categorized by their clinical diagnosis as they were discovered. These clinical categories include the recessive diseases enhanced S-cone syndrome (ESCS), Goldmann-Favre syndrome (GFS), and clumped pigmentary retinal degeneration (CPRD) [41,42,43]. NR2E3 mutations are also associated with up to 1% of all autosomal dominant retinitis pigmentosa (adRP) [44, 45]. The association of NR2E3 with several clinical phenotypes and varying modes of inheritance strongly indicates that these retinal diseases manifest on a permissive or selective genetic background and are influenced, at least in part, by genetic modifier genes [46,47,48,49]. Given the role of NHRs such as NR2E3, to modulate numerous key biological networks essential for maintaining retinal homeostasis, this study evaluated Nr2e3 as a broad-spectrum genetic modifier with the potential to attenuate retinal degeneration in several different mouse models.
In this study, the efficacy of subretinal delivery of AAV8-Nr2e3 to attenuate and ameliorate retinal degeneration was assessed in five independent RP models that represent the heterogeneity observed in human RP disease. The five RP models tested were FVB-Pde6ß rd1/NJ (rd1), Rhodopsin null allele (Rho−/−), B6.129S6(Cg)-Rhotm1.1Kpal/J (RhoP23H), BXD24/TyJ-Cep290rd16/J (rd16) and Nr2e3rd7/J (rd7) (Table 1). The rd1 mouse, representing the most severe and early form of human retinal degeneration, harbors a mutant Pde6b gene mapped on chromosome 5 [50,51,52,53,54,55]. The mutant Pde6b gene contains a murine leukemia provirus insertion in intron 1 and a point mutation, which introduces a stop codon in exon 7 (Y347STOP) [56, 57]. Independent of this, a second mutation has been found in this gene, which is the integration of a murine leukemia virus in the first intron of the 6beta (Pde6ß) gene [58]. Mutations in human PDE6ß are associated with RP and autosomal dominant congenital stationary night blindness in humans [59,60,61].
The rhodopsin null (Rho−/−) and the dominant negative RhoP23H alleles both lack a functional rhodopsin gene [62, 63]. Rho−/− mice lack expression of rhodopsin mRNA and protein [64]. In contrast, RhoP23H mice are functional nulls with an amino acid substitution of proline to histidine at position 23 that generate an aberrant message leading to protein misfolding and degradation [63]. Specifically, RhoP23H protein undergoes incomplete glycosylation and is retained in the endoplasmic reticulum (ER) and/or Golgi apparatus where it is degraded [63]. Mutations in the human rhodopsin gene account for the largest portion of inherited retinal degenerations of known genetic etiology [65]. Further, the RhoP23H mutation in particular is one of the most commonly known causes of adRP in humans [63].
The Cep290rd16 (rd16) mouse harbors a mutation in the centrosomal protein Cep290 that results in early-onset retinal degeneration with autosomal recessive inheritance [66]. Mutations in human CEP290 are associated with several syndromic and nonsyndromic forms of retinal degeneration [66, 67]. The rd7 mouse is a model for Nr2e3 associated retinal degenerations. rd7 mice, harboring a recessive mutation in Nr2e3, are clinically characterized by pan retinal spots apparent at eye opening (postnatal (P) day 14), and whorls and rosettes in the outer nuclear layer (ONL) observed histologically by P10 [68, 69]. rd7 mice have two distinct outcomes: a disruption in development of cone cells causing a significant increase of blue opsin expressing cone cells, and progressive degeneration of rod and cone photoreceptor cells [68, 70]. Results of this study show that the administration of AAV8-Nr2e3 therapy improves clinical, histological, functional, and molecular disease outcomes in each of the five models of retinal disease. These studies demonstrate the mechanism of Nr2e3 therapy involves resetting key retinal transcription factors and key biological networks that work in concert with Nr2e3 to modulate the homeostatic state of the retina. This research is predicated on the fact that disease outcome is rarely due to a single gene mutation; rather, it is a result of the combinatorial mutational load on the biological system, which is often strongly influenced by other factors such as modifier genes. This study demonstrates a novel approach to gene therapy and suggests that Nr2e3 can potentially serve as a broad-spectrum gene therapy to attenuate retinal degeneration.
Materials and methods
Power calculations to determine statistical significance and inclusion/exclusion criteria
Power calculations were conducted using G*Power 3.1 software analysis for estimating sample size required for each analysis and quantification described as indicated. Using means and standard deviation defined by previously published studies, a minimal number of seven animals per experimental group were used to provide 90% power and 30% difference at significant level of 0.05. The data have been analyzed double-blinded using two-way ANOVA comparisons. Animals with unresolved surgical trauma, premature unintended death, or cataracts were excluded from the study. Approximately 20% of the 600 total experimental animals were excluded from all analyses based on these criteria.
Animal maintenance
Animals used in this study were housed and bred in the vivarium at the Schepens Eye Research Institute under standard conditions. C57BL6/J (Jax stock #000664), FVB-Pde6ß rd1/NJ (rd1; Jax stock #001800), B6.129S6(Cg)-Rhotm1.1Kpal/J (RhoP23H; Jax stock #017628), BXD24/TyJ-Cep290rd16/J (rd16; Jax stock #000031), and Nr2e3rd7/J(rd7; Jax stock #002139) mice were obtained from Jackson Laboratories, Bar Harbor, ME. Rhodopsin Knock-out (Rho−/−) mice lacking expression of rhodopsin [59] were generously donated by Dr. C. Cepko (Harvard Medical School).
AAV8-Nr2e3 cloning and preparation
AAV8-Nr2e3 vector was generated at the Gene Transfer Vector Core, Grousbeck Gene Therapy Center, Mass Eye and Ear (http://vector.meei.harvard.edu/). Briefly, HEK293 cells were transfected with the AAV8 rep-cap packaging, Ad-helper, and AAV2 ITR-flanked transgene constructs. After 3 days, cells and media were harvested in high salt conditions, treated with Benzonase, and cellular debris was precipitated. The supernatant was subjected to tangential flow filtration and retentate was subsequently subjected to Iodixanol ultracentrifugation density gradient. AAV fractions were collected and buffer exchange was performed for final formulation in phosphate buffered solution (PBS) + 5% glycerol. The ubiquitous CAG promoter was used in the vector. CAG is a strong synthetic hybrid promoter consisting of the cytomegalovirus enhancer fused to the chicken beta-actin promoter. Mouse Nr2e3 cDNA to be packaged into AAV8 was generated by RT-PCR from mRNA of a B6 mouse retina using the following primers: forward: GCTGTACAAGGGCGGATGAGCTCTACAGTGGCT; reverse: ATACCGGTTGGCACTCCCAACTAGTT. These primers were introduced at the restriction sites BsrGI at the 5′ end and AgeI at the 3′ end of Nr2e3 cDNA, and were used for cloning into the pZac2.1-CASI-eGFP-RGB plasmid (also known as pAAV). Final products were verified by restriction enzyme digestions and sequencing.
AAV5-Nr2e3-GFP and AAV2.7m8-Nr2e3 cloning and preparation
The AAV2.7m8 plasmid was obtained from Addgene, a gift from John Flannery and David Schaffer [71] (Addgene plasmid #64839: http://n2t.net/addgene:64839;RRID:Addgene_64839) (Supplementary Fig. 1). AAV5-Nr2e3-GFP and AAV2.7m8-Nr2e3 were cloned and constructed by VectorBioLabs (Malvern, PA, USA), similar to the method described above for AAV8-Nr2e3. Mouse Nr2e3 was introduced into restriction sites NheI and KpnI of AAV5 and restriction sites EcoRI and XhoI of AAV2.7m8 using the following primers:
Forward: CCTAAGCTTATGAGCTCTACAGTGGCTGCCTCC
Reverse: ATCGAATTCGGATCCGGTACCCTAGTTTTTGAACATGTCACACAG
The final product was verified by restriction enzyme digest and sequencing.
Subretinal injection
All AAV-Nr2e3 constructs were delivered by subretinal injection. Control injections included no injection in the contralateral eye, untreated animals, and GFP only injections. Approximately 600 experimental animals were used in this study. No gender bias was observed and both males (48.94%) and females (51.06%) were used equally in the study. P0 pups were anesthetized on ice, and the eyelids were carefully opened along the eyelid fissure using a 30 gauge (G) needle. The 30G needle was then used to create a hole in the sclera adjacent to the limbus, and a blunt 33G cannula attached to a Hamilton syringe was advanced into the eye. A slight resistance to the needle indicated Bruch’s membrane was reached. A total of 1 × 109 viral genomes (vg) in a total volume of 0.5 μL was manually injected slowly and gently into the subretinal space of the adult or P0 mice. Subretinal injection was performed in adults as described above after anesthetizing animals by intraperitoneal (IP) injection with a mixture of ketamine (1 mg/mL) and xylazine (0.4 mg/mL).
Clinical examination
Fundus examination and optical coherence tomography (OCT) were performed on adult injected and uninjected animals. Animals were anesthetized with a mixture of ketamine (1 mg/mL) and xylazine (0.4 mg/mL) and pupils were dilated with 1% tropicamide. Fundus images were taken using the Micron III Retinal Imaging Camera and Stream Pix software (Phoenix Research Laboratories, Pleasanton, CA, USA). Following fundus imaging, OCT was performed using the Bioptigen OCT scanner and software. Mice were restrained in a mounting tube and the fundus camera in the optical head of the apparatus and alignment was guided by monitoring and optimizing the real time OCT image of the retina. Four rotational cross section scans (dorsal–ventral and nasal–caudal) with 100 series/scan were taken for each retina. Data was analyzed using Bioptigen OCT software (N = 10/strain/experimental group).
Electroretinography
Electroretinography (ERG) analysis was performed on Nr2e3 treated and untreated animals as described previously [72]. Briefly, mice were anesthetized with an IP injection of 1 mg/mL ketamine and 0.4 mg/mL xylazine in a saline carrier (10 mg/g of body weight), and mouse eyes were dilated with 1% tropicamide and 2.5% phenylephrine hydrochloride applied topically. Dark- and light-adapted ERGs were performed using the Espion Visual Electrophysiology System (Diagnosys, Littleton, MA) with gold loop electrodes (Diagnosys LLC) placed on the apex of the cornea. A reference needle electrode was inserted subcutaneously in the forehead and a ground electrode was placed subcutaneously at the base of the tail. For scotopic recordings, mice were dark adapted for at least 6 h and then anesthetized before recording. Dark-adapted responses were recorded to short wavelength (λmax = 470 nm; Wratten 47A filter; Kodak, Rochester, NY) flashes of light over a 4.0-log unit range of intensities (0.3-log unit steps). Light-adapted responses were obtained with white flashes (0.3 log unit steps) on a rod-saturating background after 10 min of exposure to the background light to allow complete light adaptation. Signal processing was performed using EM for Windows v7.1.2. Signals were sampled every 0.8 ms over a response window of 200 ms (LKC Technologies, Inc., Gaithersburg, MD). Responses were averaged for each stimulus condition with up to 50 records for the weakest signals. Dark-adapted responses and light-adapted responses illustrated in this study were obtained using stimulator intensities of 24.1 cd s/m2 for scotopic responses and 25.6 cd s/m2 for photopic responses (N = 7/strain/experimental group).
Histology
Following euthanasia, eyes were cauterized to mark dorsal orientation, and enucleated. Tissue samples were collected and immediately immersed in freshly made 4% paraformaldehyde in 1× PBS or in 3:1 methanol/acetic acid overnight at 4 °C. Eyes were then paraffin embedded with dorsal/ventral orientation and 5 µm sections were collected over 100 µm of retinal depth and processed for hematoxylin/eosin staining. Briefly, retina sections were deparaffinated in xylene and ethanol washed and stained with hematoxylin and eosin Y. Slides were mounted with Permount mounting medium. Over 500 μm of sections/animal were visualized and representative images captured with the Leica DMI6000 microscope. As outer plexiform layer (OPL) collapse was observed in some Nr2e3 treated retinas, the first five layers of cells were counted as inner nuclear layer (INL) and the rest of cell layers were considered as ONL when counting the ONL cell layer number. Quantification of percent observed rescue was determined by comparing treated to control B6 ONL. Cell counts were performed in a double-blinded manner over 100 μm retinal area (N = 10/strain/experimental group).
Immunohistochemistry
Immunohistochemistry analysis was performed on 10 µm paraffin embedded serial sections from the enucleated mouse eyes as described in our previous studies [72]. At minimum 100 µm of retina/sample was evaluated by IHC. Briefly, sections were blocked with 2% normal horse serum (#S-2000 VectorLabs, CA) in PBS, and incubated with the following cell type-specific primary antibodies in a 1:200 dilution: rhodopsin (mouse monoclonal, Millipore MAB5356); green/red opsin (rabbit polyclonal, Millipore AB5405); blue opsin (rabbit polyclonal, Millipore AB5407); GFP (1:500, rabbit polyclonal, Abcam ab290). The following day, sections were rinsed with PBS and incubated with the corresponding secondary antibody (1:400 Alexa fluor 488 goat antirabbit, Invitrogen A11008) and nuclei were stained with 4,6-Diamidino-2-Phenylindole, Dihydrochloride (DAPI). Over 500 μm of sections/animal were visualized and representative images of IHC labeling were captured using a Leica DMI6000 fluorescent microscope equipped with the appropriate bandpass filter for each fluorochrome. Cell counts were performed in a double-blinded manner over 100 μm retinal area (N = 10/strain/experimental group).
Retinal whole mount IHC
Retina whole mounts have been performed as previously described [73]. Microdissection of the retina for whole mounts were performed as follows: the anterior eye segments, including the iris, were removed with a round cut along the limbus using microdissection scissors. The lens was then removed and the retina was gently separated from the pigmented epithelium and the choroidal-scleral complex. Whole retinas were transferred to a 96-well culture plate and immunohistochemistry was performed as follows. Retina cups were blocked with 2% normal horse serum (#S-2000 VectorLabs, CA) in PBS with 0.02% sodium azide, 1% bovine serum albumin (BSA), and 0.1% Triton X 100; and then incubated with the following cell type-specific primary antibodies in a 1:100 dilution: rhodopsin (mouse monoclonal, Millipore MAB5356), green opsin (rabbit polyclonal, Millipore AB5405) and blue opsin (rabbit polyclonal, Millipore AB5407) overnight, and incubated with the corresponding secondary antibody (1:200 Alexa fluor, Invitrogen) overnight. Retinas were flowered with radial incisions: two horizontal and two vertical incisions at 3, 6, 9, and 12 clock, starting from the edge toward the optic nerve and cutting 2/3 of the distance from the periphery to the center, giving the retina a cross-like form. Retinas were flat mounted on a microscope slide. Opsin labeling was visualized and images captured using a Leica DMI6000 fluorescent microscope equipped with the appropriate bandpass filter for each fluorochrome (N = 10/strain/experimental group).
Quantitative real time PCR (qRT-PCR)
Total RNA was extracted from whole retinas using the Trizol method as described previously [72]. Briefly, 2 µg of total RNA was reverse transcribed using Retroscript (Ambion AM1710) to generate cDNA. The cDNA samples were diluted 1:100 and real time PCR was performed in triplicates for each primer using Sybr Green PCR master mix (Thermo fisher #4309155). The real time PCR primers (Supplementary Tables 1 and 2) were designed using NCBI Primer-Blast and were specific for each target gene. Reactions were quantified using an ABI Step One Plus Real Time PCR and analyzed with the corresponding software. Relative expression levels were determined by normalizing cycle threshold values to the amount of β-actin expressed (1000/2Ct gene—Ct β-actin). Statistical significance of differential expression was assessed using a T-test and P value of <0.05. Results are mean ± SEM (N = 7). Primers amplifying SV40 from AAV8 vector were used to determine exogenous Nr2e3 expression (F:AGCAATAGCATCACAAATTTCACAA; R:CCAGACATGATAAGATACATTGA).
Chromatin immunoprecipitation—RT-PCR
Chromatin immunoprecipitation (chIP) was performed using P30 C57Bl6/J mouse retinas as previously described [49]. A total of 8–10 retinas were used per chIP reaction. Briefly, tissue was dissociated, homogenized, and cross-linked in 37% formaldehyde and sonicated to generate sheared fragments of 400–600 bp. Immunoprecipitation was performed overnight using 1 μg of NR2E3 antibody, goat IgG antibody served as a negative control, and the input (positive control) was not incubated with antibody. Immunoprecipitated samples were reverse cross-linked. Nr2e3 putative target genes were analyzed for nuclear receptor response element (RE) binding site using the classic (AAGTCA (n = 1–4) AAGTCA) RE binding sequence of Nr2e3 as determined algorithmically by NUBIscan124. Binding sites were searched for in a maximum of 100 kb upstream region of each gene’s start site and into intron 1. Real time primers were selected flanking putative RE sites with an average amplicon size of 200 bp (Supplementary Table 3). Quantitative RT-PCR was performed using 1 µl of 1:100 dilution (input) and 1:10 dilution (samples and immunoglobulin G (IgG) control). All sample data were normalized to IgG control. Results are mean ± SEM (N = 7/strain/experimental group).
Results
Overexpression of Nr2e3 has no detrimental effect on the retina
C57BL6/J (B6) animals were treated with AAV8-Nr2e3-GFP fusion protein to evaluate any potential detrimental effects of overexpression of Nr2e3, as well as the timing of construct expression post delivery. B6 mice were injected at P0 and evaluated at P7 and 1 month. No observable degeneration was detected in the retina post injection (Fig. 1a). Consistent with clinical findings, AAV delivery of Nr2e3 did not cause aberrant morphological changes, and immunolabeling of rod and cone opsins revealed no observable difference between injected and uninjected animals (Fig. 1a). Functional output of the retina, as detected by electroretinogram (ERG) of rod and cone responses, showed no significant differences between injected and uninjected eyes (Fig. 1b). Examination of AAV8-EGFP-Nr2e3 expression at P7 and P30 revealed that expression of the vector construct was confirmed at P30 (Fig. 1c). These results confirm that overexpression of Nr2e3 by subretinal AAV8-Nr2e3 injection is not detrimental to the retina.
B6 control AAV8-Nr2e3 treated animals show no abnormalities in a. Fundus, hematoxylin/eosin histology staining, and blue, green, and rhodopsin labeling of photoreceptor cells; and b ERG response of control B6 treated and untreated. Animals injected at P0, tissue collected at P30. c GFP label of AAV8-Nr2e3-GFP injected at P0, GFP expression assessed at P7 and P30. N = 5.
Vector or GFP alone do not affect the retina
Animals were injected with AAV8-EGFP at P0 and evaluated at 1 month to demonstrate that an empty vector alone is not sufficient for or contributes to the rescue observed in Nr2e3 treated animals. Immunohistochemistry analysis confirmed vector expression without any abnormal morphological changes (Fig. 2a). All models except rd7 have 0–1 cells in the ONL by P30, thus GFP is observed in other layers yet has no impact on the disease. Semiquantitative analysis of SV40 polyA gene expression shows no significance difference in expression of SV40 in AAV8-EGFP treated retinas as compared with AAV8-EGFP-Nr2e3 treated animals (Fig. 2b). ERG analysis also suggests that there is neither rescue nor any detrimental effect on functionality of the retina of all mutant strains when treated with empty vector only (AAV8-EGFP) (Fig. 2c).
a Immunohistochemistry of AAV8-GFP (rd1, Rho−/−, RhoP23H, rd16, and rd7). All RP models except rd7 have only 0–1 cells in the ONL at P30 and GFP expression is more pronounced in other layers yet has no impact on disease. b Semiquantitative analysis of SV40 (part of AAV8) expression in untreated, AAV8-GFP, and AAV8-Nr2e3 retinas of B6 control and RP models relative to beta-actin. c ERG B-wave amplitudes of uninjected and AAV8-GFP injected RP models and B6 control. Animals injected at P0, tissue evaluated at P30. ERGs recorded at P30. Results are mean ± SEM. N = 7.
AAV delivery of Nr2e3 in RP models before disease onset attenuates retinal degeneration
The ability of Nr2e3 to rescue retinal degeneration before disease onset was tested by subretinal delivery of AAV8-Nr2e3 in five mouse models of RP. All models except rd1 were injected at P0 and evaluated at 3–4 months of age. rd1 animals were injected at P0 and evaluated at 1 month of age due to their accelerated rate of disease progression. Although not all models have a clinical phenotype, considerable improvements were observed in the fundus of RhoP23H, rd16, and rd7 mice (Fig. 3). Interestingly, we observed the rd16 mice have a red fundus with increased and pronounced vessels (not previously reported). While no vascular leakage has been observed in rd16 mice when examined by fluorescein angiography (our unpublished data), the fundus observation resolves with Nr2e3 administration. Improvement was observed in the rd7 phenotype, with reduction of retinal spots in AAV8-Nr2e3 treated eyes compared with untreated eyes at 3 months post injection.
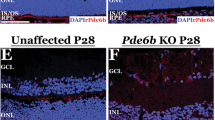
Photoreceptor degeneration often disrupts retinal topography and present with abnormal morphology. Histology analysis shows AAV8-Nr2e3 therapy improves retinal morphology and integrity in RP models. The normal mouse retina is comprised of 10–12 layers of rod and cone photoreceptor nuclei in the ONL and 5–6 layers of inner retinal cells in the INL. In the retinal degeneration mouse models evaluated, the INL did not change in nuclei number, and the ONL presented zero or only one layer of cells at the time of evaluation, with the exception of the rd7 model that presents with increased cone cells with whorls and rosettes in the ONL. Hematoxylin/eosin (H/E) staining revealed that subretinal delivery of AAV8-Nr2e3 at P0 rescued photoreceptor cells and helped maintain retinal integrity of RP retinas in all models tested at 1 month (rd1) post treatment, or 3–4 months (Rho−/−, RhoP23H, rd16, and rd7) post treatment (Fig. 4a).The attenuation of disease phenotype, as observed by ONL thickness, varied among each strain. Partial rescue (~30–80%) of the ONL count in all treated retinas was observed (Fig. 4b). It is also noteworthy that there is absolutely no gender bias in the rescue of animals as 50% of the rescued animals are male and 50% are female. Interestingly, retinal whorls and rosettes that are characteristic of the rd7 phenotype, resolved following Nr2e3 treatment, suggesting that the delivery of Nr2e3 at P0 can restore normal retinal development (Fig. 4a). Although a clear boundary between INL, ONL, and OPL was difficult to visualize in some Nr2e3 treated retinas, ONL was significantly increased in the rescued portion of treated retinas (Fig. 4b). rd1 retinas showed a profound rescue of photoreceptor cells, with a total of 6–8 nuclei layers observed in the treated eye. Rho−/−, RhoP23H, and rd16 mice showed a more moderate increase of 3–6 layers of ONL in Nr2e3 treated retinas compared with 0–1 layer in the untreated eyes of each model (Fig. 4c). Although only partial rescue was observed in all models, studies in patients have demonstrated that retention of only a single layer of photoreceptor cells can be enough to maintain minimal visual function suggesting that an increase of even 20% is significant [61, 74]. Thus, Nr2e3 therapy could have great promise in restoring retinal development.
B6, rd1, Rho−/−, RhoP23H, rd16, and rd7 animals injected at P0, evaluated at P30 (B6 and rd1) or P90-P120 (Rho−/−, RhoP23H, rd16, and rd7). a Hematoxylin/eosin staining of AAV8-Nr2e3 treated and untreated retinas with white boxes indicating location of cell count. b Rescued and un-rescued regions in retinas treated with AAV8-Nr2e3. c Cell layer numbers of ONL from AAV8-Nr2e3 treated and untreated animals in different RP models. Results are mean ± SEM. N = 7.
AAV8-Nr2e3 therapy preserves cone and rod opsin expression in five models of RP
Immunohistochemical analysis of blue and green cone opsins and rhodopsin was performed to determine if Nr2e3 therapy can restore opsin expression and thus provide a molecular reset in retinal degeneration models. Eyes from treated and untreated animals were collected at 1 month (rd1) or 3–4 months (Rho−/−, RhoP23H, rd16, and rd7) post injection and labeled with antibodies to green and blue cone opsins and to rhodopsin for rods. Untreated eyes showed no opsin-positive photoreceptor cells, except that of rd7 and Rho−/−. Consistent with our previous publication, rd7 retinas show an increase in blue opsin and slow progressive loss of all opsins over 5–16 months (Fig. 5) [70]. These studies show Rho−/− mice have sparse expression of blue and green opsin expressing cones at 1 month (Fig. 5). Interestingly, rhodopsin expression was observed in RhoP23H retinas treated with Nr2e3. The semiquantitative analysis of the blue and green opsin-positive cells shows that there is partial rescue of photoreceptor cells in rd1, Rho−/−, RhoP23H, and rd16 (Fig. 6). En face view of blue and green opsin expression in whole mount retinas of rd1, Rho−/−, RhoP23H, and rd16 Nr2e3 treated animals confirm observations of retinal sections with partial rescue of cone opsin expression in each model, consistent with H/E stain of partial rescue of ONL cells (Fig. 7, opsin expressed regions outlined by a dashed line). By 1-month age, no expression of blue or green opsin was observed in untreated rd1, RhoP23H, and rd16 animals. Similar to IHC in sections, Rho−/− retinas show sparse expression of blue and green opsin. In contrast, all treated animals showed restored blue and green opsin expression (Fig. 7). IHC performed on 1 or 3 month treated animals show consistent expression of the cone opsin genes demonstrating sustained rescue.
Improved ERG responses observed in AAV-Nr2e3 treated RP retinas
RP disease progression results in the loss of rod and cone function that is assessed by abnormal ERG responses [62, 63, 75,76,77,78,79]. Our previously published study showed improvement of ERG in rd7 mice with Nr2e3 [70]. In the present study, the visual function of Nr2e3 treated RP retinas was examined in four out of five RP strains, excluding rd7, by recording dark-adapted and light-adapted ERGs to evaluate rod- and cone-driven responses. Consistent with histology and IHC studies, partial rescue is observed in Nr2e3 treated animals compared with untreated animals (Fig. 8a). Significant percent increase of the scotopic amplitudes is observed in the treated mutant mice compared with untreated controls in each model. (Fig. 8b).
AAV8-Nr2e3 preserves retinal homeostasis in RP retinas
NHR genes such as Nr2e3 play key roles in modulating homeostasis by regulating many key biological processes and gene networks. Our prior studies and recent Ingenuity Pathway Analysis revealed that Nr2e3 regulates several key biological networks that are critical to maintaining retina homeostasis in the retina including phototransduction, cell survival, apoptosis, immunity, oxidative stress, ER stress, neuroprotection, and metabolism [49]. Representative subsets of treated animals (rd7, Rho−/−, and rd1) were evaluated for differential expression of genes that function in Nr2e3 regulated pathways. Seventy-five genes were evaluated from eight Nr2e3 modulated biological networks for Nr2e3 RE binding sites. We identified putative Nr2e3 binding sites in 19 out of 75 genes. Genes with a ≥1.5-fold variance change between the AAV8-Nr2e3 treated and untreated eyes were considered statistically significant. Consistent with the IHC results, the representative strains exhibit a significant change in gene expression of the opsin genes (Fig. 9a–c). Each strain had a unique set of genes that were differentially expressed between treated and untreated animals. Five out of eight networks were modulated by Nr2e3 treatment in each strain. As expected, the rd7 treated retinas had the greatest number of genes with differential expression in treated vs. untreated retinas (Fig. 9a). In addition, 10 genes were identified by chromatin immunoprecipitation (chIP)-RT-PCR as potential direct targets of Nr2e3 (Fig. 9d), nine of which were differentially expressed in rd7 treated retinas. Interestingly, the ER stress and cell survival factor, inositol—requiring enzyme 1 (Ire1)—is a potential direct target of Nr2e3 and is differentially expressed in all treated animals (Fig. 9a–c). Considering the unique mutational load of each model, it is not surprising that each mutant had a unique consortia of genes and networks reset by Nr2e3 treatment. Consistent with previous work demonstrating that Nr2e3 is a dual activator/repressor [39, 46,47,48,49, 80, 81], these genes were differentially modulated. These results illustrate that mutational load is modulated and balanced by transcription factors, including NHRs, for optimal cellular homeostasis [82, 83]. The upregulation of Ire1 in all treated models suggests at least one common network (ER stress and the promotion of cell survival) through which Nr2e3 modulates and resets homeostasis. Collectively, these findings show that while the specific reset varies among diseases, administration of Nr2e3 to RP diseased retinas has a positive impact in restoring the homeostatic state of the retina in the presence of disease, thus attenuating disease progression.
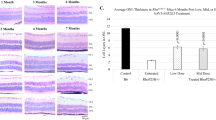
a–c Differentially expressed genes from Nr2e3-directed networks with ≥1.5-fold variance in Nr2e3 treated vs untreated retinas in rd7, rd1, and Rho−/−, respectively. Networks P phototransduction, S survival, A apoptosis, I immunity, N neuroprotection, O oxidative stress, E ER stress, M metabolic. Potential target genes of Nr2e3 are highlighted. d Potential Nr2e3 targets by Chromatin IP—real time PCR. P0 injected, samples collected at 3 months. Results are mean ± SEM. N = 7.
Studies completed by our lab and others have extensively documented the importance of Nr2e3 in photoreceptor development [69, 80, 81, 84]; however, the role of Nr2e3 in the mature retina is less understood. Recent studies reveal a key regulatory role for Nr2e3 in maintaining proper function of mature photoreceptor cells [69, 80].
The expression of Nr2e3 in all five RP mutant models was evaluated to determine if the loss of Nr2e3 contributes to RP disease. Interestingly, Nr2e3 expression in P7 (rd1) and P30 RP retinas showed a significant decrease all RP models except in the rd7−/− model. Our previous studies showed that the rd7−/− mouse, a functional null of Nr2e3, has high Nr2e3 mRNA expression but lacks protein expression [48]. These results suggest that the loss of Nr2e3 expression likely contributes to the retinal degeneration observed in each model, and the addition of Nr2e3 provides a reset for the for transcriptional signature of treated retinas (Fig. 10). Nr2e3 has been shown to function with other transcription factors such as Nr1d1, neural retinal leucine zipper (Nrl), Cone-rod homeobox (Crx), retinoic acid receptor related orphan receptor alpha (Rora), and thyroid receptor beta (Thrb) to modulate photoreceptor cell fate and retinal function as an activator or suppressor of gene expression [47,48,49, 80, 81, 85,86,87]. The expression level of five other essential retinal transcription factors (Nr1d1, Nrl, Crx, Rora, and Thrb) were determined in Nr2e3 treated and untreated retinas. Overall, a significant decrease in expression of key retinal transcription factors was reversed following Nr2e3 therapy (Fig. 10). rd1 mice lacked expression of all transcription factors tested except Crx, and these were restored with Nr2e3 therapy, significant down regulation (Fig. 10a). Both the rhodopsin models and rd16 exhibited an overall decrease in these transcription factors (Fig. 10b–d), and rd7 exhibits an overall reduction of transcription factor expression that is reset following Nr2e3 therapy (Fig. 10e).
AAV8-Nr2e3 rescues retinal degeneration after disease onset
AAV8-Nr2e3 was administered at early to intermediate stage of disease (Table 1) to determine the efficacy of Nr2e3 modifier gene therapy at a time that better represents clinical presentation. AAV8-Nr2e3 was injected subretinally at P21 and evaluated 2–3 months post injection in Rho−/−, RhoP23H, rd16, and rd7 mice. rd1 mice were injected earlier than P21 as their ONL rapidly degenerates during development. Fundus and histology show the attenuation of retinal degeneration by Nr2e3 therapy in each model (Fig. 11a, b). As shown previously, improvement varied from ~30 to 80% of the retina, depending on distribution efficiency throughout the retina. Approximately three to five layers of ONL cells were preserved in Nr2e3 treated animals compared with untreated animals that show less than or equal to one layer of ONL remaining (Fig. 11c). IHC labeling of blue and green cone opsins and rhodopsin further demonstrated the capability of Nr2e3 therapy to rescue photoreceptors after disease onset (Fig. 12). The semiquantitative analysis of the blue and green opsin-positive cells shows a partial rescue of photoreceptor cells in rd1, Rho−/−, RhoP23H, and rd16 (Fig. 13). Rhodopsin rescue is noteworthy in RhoP23H mice when treated at P0 or P21 (Figs. 5, 12, and 13), emphasizing the unique capability of Nr2e3 to modulate disease mechanism spatially and temporally.
Animals injected with AAV8-Nr2e3 at P21 and evaluated at 2–3 months post injection. a Fundus of Rho−/−, RhoP23H, rd16, and rd7. b Hematoxylin/eosin staining shows partial preservation of photoreceptor cells in treated mutant animals. c Cell layer numbers of outer nuclear layer were compared between AAV8-Nr2e3 treated and untreated animals in the four RP models and B6 control. Results are mean ± SEM. N = 7.
To confirm that the rescue observed in Nr2e3 treated retinas is not vector specific, Nr2e3 was packaged in AAV5 and AAV2.7m8 [71, 88]. Adult rd7 animals were injected with AAV5-Nr2e3-GFP or AAV2.7m8-Nr2e3 and evaluated clinically before treatment, as well as 1 month post treatment. The presence of GFP in the AAV5-Nr2e3-GFP treated retinas correlated with the region of rescue and corresponded to absence of retinal spots (Supplementary Fig. 2). Reduction of retinal spots and retinal whorls was observed in rd7 animals after 1 month of AAV5 or AAV2.7m8-delivered Nr2e3 (Supplementary Fig. 2). OCT images were scanned and captured at the same frame for the same animal before treatment and 1 month post treatment. These results demonstrate that Nr2e3 can ameliorate retinal degeneration outcomes regardless of the vehicle of delivery.
Discussion
Many factors strongly influence clinical outcome of disease including genetic modifiers (such as Nr2e3), epigenetic factors, allelic heterogeneity, and the environment [40]. Genetic modifiers play a key role in modulating disease onset, progression, and clinical outcome by either enhancing or suppressing disease [24, 40]. The discovery of genetic modifiers has provided a novel and innovative opportunity to develop powerful therapeutic strategies to treat human diseases. While tremendous work has focused on developing novel gene replacement and gene editing strategies to target mutant genes for diseases such as RP [89,90,91,92,93], it is not possible or ideal for many diseases and broader approaches must be developed. This is the first study to demonstrate successful improvement in several different mouse models of retinal degeneration by a single genetic modifier gene. Each strain showed improvement of photoreceptor survival, preservation of retinal structure, a change in gene expression, and stabilization of retinal function. While the specific reset varied between species, the cumulative impact toward the restoration of a more homeostatic state of the retina was observed. This restoration allowed for sustained impact and attenuation of RP disease.
We observed 30–80% improvement at the histological, immunohistochemical, and functional level in five RP models following treatment with AAV8-Nr2e3. AAV8 was chosen for this study due to its ability to target specificity toward photoreceptor cells. In addition, Nr2e3 expression was driven by a strong, general promoter rather than a cell type-specific promoter, as they are often not as robust. The models used in this study were chosen because they vary on severity and rate of disease progression, modeling human inherited retinal degeneration. Restoration of photoreceptor cells was consistently observed in each model by several methods. Histological analysis revealed regions of rescue in each treated eye. The rescue was demonstrated as an increase in ONL nuclei in specific regions of the retina. Similarly, IHC showed improvement of cone and rod opsin expression in regions of rescue, and it is easy to distinguish regions of rescue in retinal sections and en face views with whole mount retinas. While analysis of individual ERGs showed similar levels of improved responses in animals that showed clinical, histological, and molecular improvement, the signal to noise ratio prevented ERG analysis from confirming a robust cumulative effect by full-field ERG recordings in comparison to focal ERGs. Several factors such as dosage and variation of injection site can contribute to the variability observed in the degree of rescue per therapy treatment. In addition, the scotopic ERG response improvements in Rho−/− mice likely come from cone responses, as IHC and flat mount data demonstrate exclusive rescue of cone opsins and lack of rhodopsin expression in treated and untreated.
It is noteworthy that these studies were conducted using a low dose of AAV8-Nr2e3, 1 × 109 genome copy, which may also contribute to the modest improvement observed for some treated retinas. AAV expression has been shown to be present for months, and even years, in higher order animals, suggesting that while it is not integrated, it does provide long term stable expression [94,95,96]. The potency of Nr2e3 as a therapeutic is clearly demonstrated by its ability to attenuate retinal degeneration and restore retinal integrity in several RP models at the early to intermediate stage of the disease. In addition, our prior and current studies show that regardless of the vehicle (naked DNA, nanoparticle, AAV8, AAV5, or AAV2.7m8), Nr2e3 can rescue retinal disease in rd7.
Our prior studies demonstrated the role of Nr2e3 in modulating numerous gene networks that impact homeostasis. Here, we evaluated 75 genes from eight key Nr2e3-directed gene networks that contribute to retinal homeostasis to determine the mechanism by which Nr2e3 achieved such broad-spectrum rescue. Evaluating genes in 8 of the Nr2e3-directed gene networks revealed that improved outcome coincides with differential expression of genes in the Nr2e3-directed networks. These gene networks are key regulators maintaining the homeostatic state of the retina. While each mutant strain is directed by a unique transcriptional profile, common networks such as phototransduction, apoptosis, immunity, oxidative stress, and ER stress genes were impacted in each treated model
One biological regulatory network influenced by Nr2e3 is ER stress. For example, Ire1, a putative direct target of Nr2e3, is upregulated in treated eyes of all models tested. Ire1 encodes an ER-resident transmembrane protein that controls the most highly conserved unfolded protein response (UPR) signaling pathway during ER stress [97, 98]. Ire1, a potential gene target of Nr2e3, is upregulated in all Nr2e3 treated eyes compared with their respective controls. The ER is an essential organelle for multiple cellular functions including maintaining calcium homeostasis and the biosynthesis of proteins and lipids. Protein misfolding and the consequential accumulation of unfolded proteins in the lumen of the ER initiate the UPR to alleviate stress [99]. Ire1 undergoes oligomerization and activation of its kinase and RNase functions in response to these protein misfolding in the ER, initiating the nonconventional splicing of Xbp-1 mRNA [100]. Thereby, Ire1 signaling enhances the protein-folding environment of the ER by expanding the amount of ER and its constituent protein-folding machinery, as well as clearing misfolded ER proteins [101]. Ire1 signaling pathway transcriptionally upregulates numerous ER chaperones that were shown to promote P23H rhodopsin exit from the ER by partially ameliorating this misfolding defect [102,103,104]. The RhoP23H point mutation encodes a misfolded rhodopsin protein, one which undergoes incomplete glycosylation and is retained in the ER and/or Golgi where most of it is degraded [63, 65]. We hypothesize Ire1 upregulation in Nr2e3 treated animals enhances UPR signaling activation, allowing the P23H rhodopsin protein to escape degradation in the ER and migrate to the outer segment of the retina as observed by IHC. However, whether or not the rescued rhodopsin in RhoP23H mice upholds functional qualities has yet to be confirmed. Future studies include evaluating the role of Ire1 in RP disease. Interestingly, when RhoP23H mice were treated at P21, rhodopsin expression is not observed; however, blue and green opsin expression was rescued with AAV8-Nr2e3 delivery at P0 or P21. Therefore, treating RhoP23H with AAV8-Nr2e3 at P21 may only rescue their cones. This underscores the potential of Nr2e3 as a unique therapeutic agent to regulate complex genetic mechanisms in temporal and spatial manner.
Interestingly, our real time PCR results revealed that RP models showed very low to no expression of Nr2e3 and other key transcription factors evaluated in this study (Nrl, Crx, Trb, Rora, and Nr1d1), which likely contributed to the progression of retinal degeneration in each model. Further, Nr2e3 therapy increased the expression of these key transcription factors. This data suggest that Nr2e3 action to restore photoreceptor development and improve retinal function occurs through the modulation of many gene networks and the recruitment of key photoreceptor transcription factors that work in concert to reset retinal homeostasis.
In summary, this study demonstrates the potency of a novel modifier gene therapy to elicit broad-spectrum therapeutic effects. Modifier genes, particularly NHRs that are capable of regulating numerous networks, can attenuate disease progression and strongly impact disease outcome. The results of this study illustrate that the attenuation of disease outcomes caused by mutations in different genes can be achieved with a single modifier gene. Future studies include in-depth analysis of photoreceptor function at the onset of disease for each model and at specific time points post treatment with Nr2e3. Future studies will also evaluate promoter specificity and potency, dosage studies, and combination therapies to determine optimal efficacy. Recent reports discuss the toxicity of using strong promoters such as CMV. However, this present study employs a CAG promoter and a dose fivefold less than the lowest dose evaluated recent studies [105, 106]. The results from this study can be translated to other human diseases and lay the foundation for the development of modifier gene therapy for other neurological diseases.
References
Russell S, Bennett J, Wellman JA, Chung DC, Yu ZF, Tillman A, et al. Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: a randomised, controlled, open-label, phase 3 trial. Lancet. 2017;390:849–60.
Braun CJ, Boztug K, Paruzynski A, Witzel M, Schwarzer A, Rothe M, et al. Gene therapy for Wiskott-Aldrich syndrome-long-term efficacy and genotoxicity. Sci Transl Med. 2014;7:357–65.
Hacein-Bey-Abina S, Pai S-Y, Gaspar HB, Armant M, Berry CC, Blanche S, et al. A modified γ-retrovirus vector for X-linked severe combined immunodeficiency. N Engl J Med. 2014;371:1407–17.
Bainbridge JWB, Smith AJ, Barker SS, Robbie S, Henderson R, Balaggan K, et al. Effect of gene therapy on visual function in Leber’s congenital amaurosis. N Engl J Med. 2008;358:2231–9.
Weleber RG, Pennesi ME, Wilson DJ, Kaushal S, Erker LR, Jensen L, et al. Results at 2 years after gene therapy for RPE65-deficient leber congenital amaurosis and severe early-childhood-onset retinal dystrophy. Ophthalmology. 2016;123:1606–20.
Palfi S, Gurruchaga JM, Scott Ralph G, Lepetit H, Lavisse S, Buttery PC, et al. Long-term safety and tolerability of ProSavin, a lentiviral vector-based gene therapy for Parkinson’s disease: A dose escalation, open-label, phase 1/2 trial. Lancet. 2014;383:1138–46.
Hwu WL, Muramatsu SI, Tseng SH, Tzen KY, Lee NC, Chien YH, et al. Gene therapy for aromatic L-amino acid decarboxylase deficiency. Sci Transl Med. 2012;4:134ra61.
Marlhens F, Bareil C, Griffoin JM, Zrenner E, Amalric P, Eliaou C, et al. Mutations in RPE65 cause leber’s congenital amaurosis. Nat Genet. 1997;17:139–41.
Acland GM, Aguirre GD, Ray J, Zhang Q, Aleman TS, Cideciyan AV, et al. Gene therapy restores vision in a canine model of childhood blindness. Nat Genet. 2001;12:1072–82.
MacLaren RE, Groppe M, Barnard AR, Cottriall CL, Tolmachova T, Seymour L, et al. Retinal gene therapy in patients with choroideremia: initial findings from a phase 1/2 clinical trial. Lancet. 2014;383:1129–37.
Lenis TL, Sarfare S, Jiang Z, Lloyd MB, Bok D, Radu RA. Complement modulation in the retinal pigment epithelium rescues photoreceptor degeneration in a mouse model of Stargardt disease. Proc Natl Acad Sci. 2017;114:3987–92.
Ghazi NG, Abboud EB, Nowilaty SR, Alkuraya H, Alhommadi A, Cai H, et al. Treatment of retinitis pigmentosa due to MERTK mutations by ocular subretinal injection of adeno-associated virus gene vector: results of a phase I trial. Hum Genet. 2016;135:327–43.
Kahle NA, Peters T, Zobor D, Kuehlewein L, Kohl S, Zhour A, et al. Development of methodology and study protocol: safety and efficacy of a single subretinal injection of rAAV.hCNGA3 in patients with CNGA3—linked achromatopsia investigated in an exploratory dose-escalation trial. Hum Gene Ther Clin Dev. 2018;29:121–31.
Parmeggiani F, Sorrentino FS, Ponzin D, Barbaro V, Ferrari S, Di Iorio E. Retinitis Pigmentosa: Genes and disease mechanisms. Curr Genom. 2011;12:238–49.
Daiger SP, Sullivan LS, Bowne SJ. Genes and mutations causing retinitis pigmentosa. Clin Genet. 2013;84:132–41.
Bunker C, Berson E, Bromley W, Hayes R, Roderick T. Prevalence of retinitis pigmentosa in Maine. Am J Ophthalmol. 1984;97:357–65.
Grøndahl J. Estimation of prognosis and prevalence of retinitis pigmentosa and Usher syndrome in Norway. Clin Genet. 1987;31:255–64.
Pierrottet CO, Zuntini M, Digiuni M, Bazzanella I, Ferri P, Paderni R, et al. Syndromic and non-syndromic forms of retinitis pigmentosa: a comprehensive Italian clinical and molecular study reveals new mutations. Genet Mol Res. 2014;13:8815–33.
Ali MU, Rahman MSU, Cao J, Yuan PX. Genetic characterization and disease mechanism of retinitis pigmentosa; current scenario. 3 Biotech. 2017;7:251.
Al-maghtheh M, Inglehearn CF, Jeffrey TK, Evans K, Moore AT, Jay M, et al. Identification of a sixth locus for autosomal dominant retinitis pigmentosa on chromosome 19. Hum Mol Genet. 1994;3:351–4.
Andréasson S, Ponjavic V, Abrahamson M, Ehinger B, Wu W, Fujita R, et al. Phenotypes in three Swedish families with X-linked retinitis pigmentosa caused by different mutations in the RPGR gene. Am J Ophthalmol. 1997;124:95–102.
Blanton SH, Heckenlively JR, Cottingham AW, Friedman J, Sadler LA, Wagner M, et al. Linkage mapping of autosomal dominant retinitis pigmentosa (RP1) to the pericentric region of human chromosome 8. Genomics. 1991;11:857–69.
Hamel C, Hartong DT, Berson EL, Dryja TP, Hamel C. Retinitis pigmentosa. Orphanet J Rare Dis. 2006;1:40.
Houlston RS, Tomlinson IP. Modifier genes in humans: strategies for identification. Eur J Hum Genet. 1998;6:80–8.
Harper AR, Nayee S, Topol EJ. Protective alleles and modifier variants in human health and disease. Nat Rev Genet. 2015;16:689–701.
Chow CY, Kelsey KJP, Wolfner MF, Clark AG. Candidate genetic modifiers of retinitis pigmentosa identified by exploiting natural variation in Drosophila. Hum Mol Genet. 2016;25:651–9.
Haider NB, Ikeda A, Naggert JK, Nishina PM. Genetic modifiers of vision and hearing. Hum Mol Genet. 2002;11:1195–206.
Salvatore F, Scudiero O, Castaldo G. Genotype-phenotype correlation in cystic fibrosis: the role of modifier genes. Am J Med Genet. 2002;111:88–95.
Dipple KM, McCabe ERB. Modifier genes convert “simple” Mendelian disorders to complex traits. Mol Genet Metab. 2000;71:43–50.
Hsieh CS, Macatonia SE, Garra A, Murphy KM. T cell genetic background determines default T helper phenotype development in vitro. J Exp Med. 1995;181:713 LP–721.
Kiesewetter S, Macek M, Davis C, Curristin SM, Chu CS, Graham C, et al. A mutation in CFTR produces different phenotypes depending on chromosomal background. Nat Genet. 1993;5:274–8.
Rose-Hellekant TA, Gilchrist K, Sandgren EP. Strain background alters mammary gland lesion phenotype in transforming growth factor-α transgenic mice. Am J Pathol. 2002;161:1439–47.
Calhoun JD, Hawkins NA, Zachwieja NJ, Kearney JA. Cacna1g is a genetic modifier of epilepsy in a mouse model of Dravet syndrome. Epilepsia. 2017;58:e111–5.
Eshraghi M, McFall E, Gibeault S, Kothary R. Effect of genetic background on the phenotype of the Smn 2B/-mouse model of spinal muscular atrophy. Hum Mol Genet. 2016;25:ddw278.
Tanabe LM, Martin C, Dauer WT. Genetic background modulates the phenotype of a mouse model of dyt1 dystonia. PLoS ONE. 2012;7:e32245.
Ebermann I, Phillips JB, Liebau MC, Koenekoop RK, Schermer B, Lopez I, et al. PDZD7 is a modifier of retinal disease and a contributor to digenic Usher syndrome. J Clin Investig. 2010;120:1812–23.
Maddox DM, Ikeda S, Ikeda A, Zhang W, Krebs MP, Nishina PM, et al. An allele of microtubule-associated protein 1A (Mtap1a) reduces photoreceptor degeneration in Tulp1 and tub mutant mice. Investig Ophthalmol Vis Sci. 2012;53:1663–9.
Fernandez-Funez P, Nino-Rosales ML, De Gouyon B, She WC, Luchak JM, Martinez P, et al. Identification of genes that modify ataxin-1-induced neurodegeneration. Nature. 2000;408:101–6.
Haider NB, Zhang W, Hurd R, Ikeda A, Nystuen AM, Naggert JK, et al. Mapping of genetic modifiers of Nr2e3rd7/rd7 that suppress retinal degeneration and restore blue cone cells to normal quantity. Mamm Genome. 2008;19:145–54.
Cruz NM, Yuan Y, Leehy BD, Baid R, Kompella U, DeAngelis MM, et al. Modifier genes as therapeutics: the nuclear hormone receptor rev erb alpha (Nr1d1) rescues Nr2e3 associated retinal disease. PLoS ONE. 2014;9:e87942.
Haider N, Jacobson S, Cideciyan A, Swiderski R, Streb L, Searby C, et al. Mutation of a nuclear receptor gene, NR2E3, causes enhanced S cone syndrome, a disorder of retinal cell fate. Nat Genet. 2000;24:127–31.
Schorderet DF, Escher P. NR2E3 mutations in enhanced S-cone sensitivity syndrome (ESCS), Goldmann-Favre syndrome (GFS), clumped pigmentary retinal degeneration (CPRD), and retinitis pigmentosa (RP). Hum Mutat. 2009;30:1475–85.
Sharon D, Sandberg MA, Caruso RC, Berson EL, Dryja TP. Shared mutations in NR2E3 in enhanced S-cone syndrome, Goldmann-Favre syndrome, and many cases of clumped pigmentary retinal degeneration. Arch Ophthalmol. 2003;121:1316–23.
Coppieters F, Leroy BP, Beysen D, Hellemans J, De Bosscher K, Haegeman G, et al. Recurrent mutation in the first zinc finger of the orphan nuclear receptor NR2E3 causes autosomal dominant retinitis pigmentosa. Am J Hum Genet. 2007;81:147–57.
Gire A, Sullivan L, Bowne S, Birch D, Hughbanks-Wheaton D, Heckenlively JR, et al. The Gly56Arg mutation in NR2E3 accounts for 1-2% of autosomal dominant retinitis pigmentosa. Mol Vis. 2007;13:1970–5.
Webber AL, Hodor P, Thut CJ, Vogt TF, Zhang T, Holder DJ, et al. Dual role of Nr2e3 in photoreceptor development and maintenance. Exp Eye Res. 2008;87:35–48.
Cheng H, Aleman TS, Cideciyan AV, Khanna R, Jacobson SG, Swaroop A. In vivo function of the orphan nuclear receptor NR2E3 in establishing photoreceptor identity during mammalian retinal development. Hum Mol Genet. 2006;15:2588–602.
Haider NB, Demarco P, Nystuen AM, Huang X, Smith RS, Mccall MA, et al. The transcription factor Nr2e3 functions in retinal progenitors to suppress cone cell generation. Vis Neurosci. 2006;23:917–29.
Olivares AM, Jelcick AS, Reinecke J, Leehy B, Haider A, Morrison MA, et al. Multimodal regulation orchestrates normal and complex disease states in the retina. Sci Rep. 2017;7:690–706.
Sidman RL, Green MC. Retinal degeneration in the mouse: location of the rd locus in linkage group xvii. J Hered. 1965;56:23–9.
Lolley RN, Farber DB, Rayborn ME, Hollyfield JG. Cyclic gmp accumulation causes degeneration of photoreceptor cells: simulation of an inherited disease. Science (80-). 1977. https://doi.org/10.1126/science.193183.
Farber DB. From mice to men: the cyclic GMP phosphodiesterase gene in vision and disease: the proctor lecture. Investig Ophthalmol Vis Sci. 1995;36:263–75.
Jones BW, et al. Retinal remodeling triggered by photoreceptor degenerations. J Comp Neurol. 2003. https://doi.org/10.1002/cne.10703.
Peng YW, Hao Y, Petters RM, Wong F. Ectopic synaptogenesis in the mammalian retina caused by rod photoreceptor-specific mutations. Nat Neurosci. 2000. https://doi.org/10.1038/80639.
Strettoi E, Porciatti V, Falsini B, Pignatelli V, Rossi C. Morphological and functional abnormalities in the inner retina of the rd/rd mouse. J. Neurosci. 2002. https://doi.org/10.1523/jneurosci.22-13-05492.2002.
Bowes C, Li T, Frankel WN, Danciger M, Coffin JM, Applebury ML, et al. Localization of a retroviral element within the rd gene coding for the β subunit of cGMP phosphodiesterase. Proc Natl Acad Sci USA. 1993;90:2955–9.
Pittler SJ, Baehr W. Identification of a nonsense mutation in the rod photoreceptor cGMP phosphodiesterase β-subunit gene of the rd mouse. Proc Natl Acad Sci USA. 1991;88:8322–6
Bowes C, Li T, Danciger M, Baxter LC, Applebury ML, Farber DB. Retinal degeneration in the rd mouse is caused by a defect in the beta subunit of rod cGMP-phosphodiesterase. Nature. 1990;347:677–80.
Danciger M, Blaney J, Gao Yq, Zhao Dy, Heckenlively Jr, Jacobson SG, et al. Mutations in the PDE6B gene in autosomal recessive retinitis pigmentosa. Genomics. 1995;30:1–7.
Gopalakrishna KN, Boyd K, Artemyev NO. Mechanisms of mutant PDE6 proteins underlying retinal diseases. Cell Signal. 2017;37:74–80.
McLaughlin ME, Ehrhart TL, Berson EL, Dryja TP. Mutation spectrum of the gene encoding the beta subunit of rod phosphodiesterase among patients with autosomal recessive retinitis pigmentosa. Proc Natl Acad Sci USA. 1995;92:3249–53.
Humphries MM, Rancourt D, Farrar GJ, Kenna P, Hazel M, Bush RA, et al. Retinopathy induced in mice by targeted disruption of the rhodopsin gene. Nat Genet. 1997;15:216–9.
Sakami S, Maeda T, Bereta G, Okano K, Golczak M, Sumaroka A, et al. Probing mechanisms of photoreceptor degeneration in a new mouse model of the common form of autosomal dominant retinitis pigmentosa due to P23H opsin mutations. J Biol Chem. 2011;286:10551–67.
Lem J, Krasnoperova NV, Calvert PD, Kosaras B, Cameron DA, Nicolò M, et al. Morphological, physiological, and biochemical changes in rhodopsin knockout mice. Proc Natl Acad Sci USA. 1999;96:736–41.
Shokravi MT, Dryja TP. Retinitis pigmentosa and the rhodopsin gene. Int Ophthalmol Clin. 1993;33:219–28.
Chang B, Khanna H, Hawes N, Jimeno D, He S, Lillo C, et al. In-frame deletion in a novel centrosomal/ciliary protein CEP290/NPHP6 perturbs its interaction with RPGR and results in early-onset retinal degeneration in the rd16 mouse. Hum Mol Genet. 2006;15:1847–57.
Shen T, Guan L, Li S, Zhang J, Xiao X, Jiang H, et al. Mutation analysis of Leber congenital amaurosis-associated genes in patients with retinitis pigmentosa. Mol Med Rep. 2015;11:1827–32.
Akhmedov NB, Piriev NI, Chang B, Rapoport AL, Hawes NL, Nishina PM, et al. A deletion in a photoreceptor-specific nuclear receptor mRNA causes retinal degeneration in the rd7 mouse. Proc Natl Acad Sci USA. 2000;97:5551–6.
Haider N, Naggert J, Nishina P. Excess cone cell proliferation due to lack of a functional NR2E3 causes retinal dysplasia and degeneration in rd7/rd7 mice. Hum Mol Genet. 2001;10:1619–26.
Haider NB, Naggert JK, Nishina PM. Excess cone cell proliferation due to lack of a functional NR2E3 causes retinal dysplasia and degeneration in rd7/rd7 mice. Hum Mol Genet. 2001;10:1619–26.
Dalkara D, Byrne LC, Klimczak RR, Visel M, Yin L, Merigan WH, et al. In vivo-directed evolution of a new adeno-associated virus for therapeutic outer retinal gene delivery from the vitreous. Sci Transl Med. 2013;5:189ra76–189ra76.
Olivares AM, Han Y, Soto D, Flattery K, Marini J, Molemma N, et al. The nuclear hormone receptor gene Nr2c1 (Tr2) is a critical regulator of early retina cell patterning. Dev Biol. 2017;429:343–55.
Jelcick AS, Yuan Y, Leehy BD, Cox LC, Silveira AC, Qiu F, et al. Genetic variations strongly influence phenotypic outcome in the mouse retina. PLoS ONE. 2011;6:e21858.
Li Z-Y, Jacobson SG, Milam AH. Autosomal dominant retinitis pigmentosa caused by the threonine-17-methionine rhodopsin mutation: retinal histopathology and immunocytochemistry. Exp Eye Res. 1994;58:397–408.
Chang B, Hawes NL, Pardue MT, German AM, Hurd RE, Davisson MT, et al. Two mouse retinal degenerations caused by missense mutations in the β-subunit of rod cGMP phosphodiesterase gene. Vis Res. 2007;47:624–33.
Xu J, Dodd RL, Makino CL, Simon MI, Baylor DA, Chen J. Prolonged photoresponses in transgenic mouse rods lacking arrestin. Nature. 1997;389:505–9.
Farber DB, Flannery JG, Bowes-Rickman C. The rd mouse story: 70 years of research on an animal model of inherited retinal degeneration. Prog Retinal Eye Res. 1994;13:31–64.
Cheng H, Khan NW, Roger JE, Swaroop A. Excess cones in the retinal degeneration rd7 mouse, caused by the loss of function of orphan nuclear receptor Nr2e3, originate from early-born photoreceptor precursors. Hum Mol Genet. 2011;20:4102–15.
Gibson R, Fletcher EL, Vingrys AJ, Zhu Y, Vessey KA, Kalloniatis M. Functional and neurochemical development in the normal and degenerating mouse retina. J Comp Neurol. 2013;521:1251–67.
Cheng H, Khanna H, Oh ECT, Hicks D, Mitton KP, Swaroop A. Photoreceptor-specific nuclear receptor NR2E3 functions as a transcriptional activator in rod photoreceptors. Hum Mol Genet. 2004;13:1563–75.
Peng G, Ahmad O, Ahmad F, Liu J, Chen S. The photoreceptor-specific nuclear receptor Nr2e3 interacts with Crx and exerts opposing effects on the transcription of rod versus cone genes. Human molecular genetics. Hum Mol Genet. 2005;14:747–64.
Olivares AM, Moreno-Ramos OA, Haider NB. Role of nuclear receptors in central nervous system development and associated diseases. J Exp Neurosci. 2015;9:S93–121.
Repa JJ, Mangelsdorf DJ. The role of orphan nuclear receptors in the regulation of cholesterol homeostasis. Annu Rev Cell Dev Biol. 2000;16:459–81.
Haider N, Mollema N, Gaule M, Yuan Y, Sachs A, Nystuen A, et al. Nr2e3-directed transcriptional regulation of genes involved in photoreceptor development and cell-type specific phototransduction. Exp Eye Res. 2009;89:365–72.
Nakamura PA, Tang S, Shimchuk AA, Ding S, Reh TA. Potential of small molecule–mediated reprogramming of rod photoreceptors to treat retinitis pigmentosa. Investig Ophthalmol Vis Sci. 2016;57:6407–15.
Webber A, Hodor P, Thut C, Petrukhin K. Dual role of Nr2e3 in photoreceptor development and maintenance. Exp Eye Res. 2008;87:35–48.
Haider NB, Zhang W, Hurd R, Ikeda A, Nystuen AM, Naggert JK, et al. Mapping of genetic modifiers of Nr2e3 rd7/rd7 that suppress retinal degeneration and restore blue cone cells to normal quantity. Mamm Genome. 2008;19:145–54.
Charbel Issa P, De Silva SR, Lipinski DM, Singh MS, Mouravlev A, You Q, et al. Assessment of tropism and effectiveness of new primate-derived hybrid recombinant AAV serotypes in the mouse and primate retina. PLoS ONE. 2013;8:e60361.
Beltran WA, Cideciyan AV, Lewin AS, Iwabe S, Khanna H, Sumaroka A, et al. Gene therapy rescues photoreceptor blindness in dogs and paves the way for treating human X-linked retinitis pigmentosa. Proc Natl Acad Sci USA. 2012;109:2132–7.
Dejneka NS, Surace EM, Aleman TS, Cideciyan AV, Lyubarsky A, Savchenko A, et al. In utero gene therapy rescues vision in a murine model of congenital blindness. Mol Ther. 2004;9:182–8.
Cai X, Conley SM, Nash Z, Fliesler SJ, Cooper MJ, Naash MI. Gene delivery to mitotic and postmitotic photoreceptors via compacted DNA nanoparticles results in improved phenotype in a mouse model of retinitis pigmentosa. FASEB J. 2010;24:1178–91.
Xiao-Jie L, Hui-Ying X, Zun-Ping K, Jin-Lian C, Li-Juan J. CRISPR-Cas9: a new and promising player in gene therapy. J Med Genet. 2015;52:289–96.
Caplen NJ. Gene therapy progress and prospects. Downregulating gene expression: the impact of RNA interference. Gene Ther. 2004;11:1241.
Vandenberghe LH, Bell P, Maguire AM, Cearley CN, Xiao R, Calcedo R, et al. Dosage thresholds for AAV2 and AAV8 photoreceptor gene therapy in monkey. Sci Transl Med. 2011;3:88ra54.
Guziewicz KE, Zangerl B, Komáromy AM, Iwabe S, Chiodo VA, Boye SL, et al. Recombinant AAV-mediated BEST1 transfer to the retinal pigment epithelium: analysis of serotype-dependent retinal effects. PLoS ONE. 2013;8:e75666.
Acland GM, Aguirre GD, Bennett J, Aleman TS, Cideciyan AV, Bennicelli J, et al. Long-term restoration of rod and cone vision by single dose rAAV-mediated gene transfer to the retina in a canine model of childhood blindness. Mol Ther. 2005;12:1072–82.
Hetz C, Glimcher LH. Fine-tuning of the unfolded protein response: assembling the IRE1alpha interactome. Mol Cell. 2009;35:551–61.
Alavi MV, Chiang WC, Kroeger H, Yasumura D, Matthes MT, Iwawaki T, et al. In vivo visualization of endoplasmic reticulum stress in the retina using the ERAI reporter mouse. Investig Ophthalmol Vis Sci. 2015;56:6961–70.
Athanasiou D, Aguilà M, Bevilacqua D, Novoselov SS, Parfitt DA, Cheetham ME. The cell stress machinery and retinal degeneration. FEBS Lett. 2013;587:2008–17.
Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–91.
Chiang W-C, Messah C, Lin JH. IRE1 directs proteasomal and lysosomal degradation of misfolded rhodopsin. Mol Biol Cell. 2012;23:758–70.
Noorwez SM, Kuksa V, Imanishi Y, Zhu L, Filipek S, Palczewski K, et al. Pharmacological chaperone-mediated in vivo folding and stabilization of the P23H-opsin mutant associated with autosomal dominant retinitis pigmentosa. J Biol Chem. 2003;278:14442–50.
Noorwez SM, Malhotra R, McDowell JH, Smith KA, Krebs MP, Kaushal S. Retinoids assist the cellular folding of the autosomal dominant retinitis pigmentosa opsin mutant P23H. J Biol Chem. 2004;279:16278–84.
Noorwez SM, Sama RRK, Kaushal S. Calnexin improves the folding efficiency of mutant rhodopsin in the presence of pharmacological chaperone 11-cis-Retinal. J Biol Chem. 2009;284:33333–42.
Khabou H, Cordeau C, Pacot L, Fisson S, Dalkara D. Dosage THresholds and Influence of Transgene Cassette in Adeno-associated Virus–related Toxicity. Hum Gene Ther. 2018;29:1235–41.
Xiong W, Wu DM, Xue Y, Wang SK, Chung MJ, Ji X, et al. AAV cis-regulatory sequences are correlated with ocular toxicity. Proc Natl Acad Sci. 2019;116:5785–94.
Sakami S, Kolesnikov AV, Kefalov VJ, Palczewski K. P23H opsin knock-in mice reveal a novel step in retinal rod disc morphogenesis. Hum Mol Genet. 2014;23:1723–41.
Acknowledgements
We thank Sophie Nystuen and Sophia Snyder for technical assistance. We thank the Gene Transfer Vector Core and the SERI-MEE Morphology and Microscopy Core for their services, partly subsidized by Research to Prevent Blindness.
Funding
This work was supported by the following funding agencies: NIH-NEI RO1EY017653 (NBH), NIH-NEI RO1EY017653-01A2S1 (NBH), NIH-NEI R01EY10542 (WBT), Ocugen INC, an unrestricted grant from Research to Prevent Blindness, Inc., New York, NY to Department of Ophthalmology, Harvard Medical School (NBH), Senior Scientific Investigator Award from Research to Prevent Blindness (WBT), Hope for Vision (NBH), Massachusetts Lions Eye Research Fund (NBH), American Macular Degeneration Foundation (NBH), Webster Foundation (NBH). Research reported in this publication was supported by the National Eye Institute of the National Institutes of Health under Award Number P30EY003790.
Author information
Authors and Affiliations
Contributions
Conceived and design the study: NBH. Conducted the experiments: SL, ZL, EB, SD, KF, JC, KY, SW, MI, AU. Analyzed the data: NBH, WBT, MMD, SL, VW, SD, EB, and ZL. Wrote and edited the paper: SL, SD, ZL, EB, VW, MMD, WBT, and NBH. All authors reviewed the paper.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institute of Health. Animal use and procedures were approved by the Schepens Eye Research Institute Animal Care and Use Committee (Protocol Number: S485-0620) in compliance with the Animal Welfare Act Regulations. All efforts were made to minimize animal suffering.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Li, S., Datta, S., Brabbit, E. et al. Nr2e3 is a genetic modifier that rescues retinal degeneration and promotes homeostasis in multiple models of retinitis pigmentosa. Gene Ther 28, 223–241 (2021). https://doi.org/10.1038/s41434-020-0134-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41434-020-0134-z
This article is cited by
-
Preclinical dose response study shows NR2E3 can attenuate retinal degeneration in the retinitis pigmentosa mouse model RhoP23H+/−
Gene Therapy (2024)
-
Biliverdin regulates NR2E3 and zebrafish retinal photoreceptor development
Scientific Reports (2022)
-
CRISPR-Based Genome Editing as a New Therapeutic Tool in Retinal Diseases
Molecular Biotechnology (2021)
-
Genetic modifiers and phenotypic variability in neuromuscular disorders
Journal of Applied Genetics (2020)