Abstract
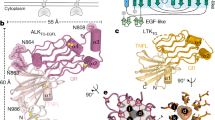
Anaplastic lymphoma kinase (ALK) is a receptor tyrosine kinase (RTK) that regulates important functions in the central nervous system1,2. The ALK gene is a hotspot for chromosomal translocation events that result in several fusion proteins that cause a variety of human malignancies3. Somatic and germline gain-of-function mutations in ALK were identified in paediatric neuroblastoma4,5,6,7. ALK is composed of an extracellular region (ECR), a single transmembrane helix and an intracellular tyrosine kinase domain8,9. ALK is activated by the binding of ALKAL1 and ALKAL2 ligands10,11,12,13,14 to its ECR, but the lack of structural information for the ALK-ECR or for ALKAL ligands has limited our understanding of ALK activation. Here we used cryo-electron microscopy, nuclear magnetic resonance and X-ray crystallography to determine the atomic details of human ALK dimerization and activation by ALKAL1 and ALKAL2. Our data reveal a mechanism of RTK activation that allows dimerization by either dimeric (ALKAL2) or monomeric (ALKAL1) ligands. This mechanism is underpinned by an unusual architecture of the receptor–ligand complex. The ALK-ECR undergoes a pronounced ligand-induced rearrangement and adopts an orientation parallel to the membrane surface. This orientation is further stabilized by an interaction between the ligand and the membrane. Our findings highlight the diversity in RTK oligomerization and activation mechanisms.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
Atomic coordinates have been deposited in the Protein Data Bank (PDB) with accession codes 7N00 (cryoEM structure of ALK-ECRABR–ALKAL2), 7MZY (X-ray structure of ALK-ECRABR(ΔEGF)), 7MZW (NMR structure of ALK-ECRABR), 7MZX (NMR structure of ALKAL2) and 7MZZ (NMR structure of ALKAL1). The cryo-EM density map of ALK-ECR–ALKAL2 has been deposited in the Electron Microscopy Data Bank (EMDB) under the accession code EMD-24095. NMR resonance assignments have been deposited in the Biological Magnetic Resonance Bank (BMRB) under the accession codes 30910 (ALK-ECRABR), 30911 (ALKAL2) and 30912 (ALKAL1).
References
Morris, S. W. et al. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin’s lymphoma. Science 263, 1281–1284 (1994).
Orthofer, M. et al. Identification of ALK in thinness. Cell 181, 1246–1262 (2020).
Hallberg, B. & Palmer, R. H. Mechanistic insight into ALK receptor tyrosine kinase in human cancer biology. Nat. Rev. Cancer 13, 685–700 (2013).
Chen, Y. et al. Oncogenic mutations of ALK kinase in neuroblastoma. Nature 455, 971–974 (2008).
George, R. E. et al. Activating mutations in ALK provide a therapeutic target in neuroblastoma. Nature 455, 975–978 (2008).
Janoueix-Lerosey, I. et al. Somatic and germline activating mutations of the ALK kinase receptor in neuroblastoma. Nature 455, 967–970 (2008).
Mosse, Y. P. et al. Identification of ALK as a major familial neuroblastoma predisposition gene. Nature 455, 930–935 (2008).
Morris, S. W. et al. ALK, the chromosome 2 gene locus altered by the t(2;5) in non-Hodgkin’s lymphoma, encodes a novel neural receptor tyrosine kinase that is highly related to leukocyte tyrosine kinase (LTK). Oncogene 14, 2175–2188 (1997).
Lemmon, M. A. & Schlessinger, J. Cell signaling by receptor tyrosine kinases. Cell 141, 1117–1134 (2010).
Zhang, H. et al. Deorphanization of the human leukocyte tyrosine kinase (LTK) receptor by a signaling screen of the extracellular proteome. Proc. Natl Acad. Sci. USA 111, 15741–15745 (2014).
Guan, J. et al. FAM150A and FAM150B are activating ligands for anaplastic lymphoma kinase. eLife 4, e09811 (2015).
Reshetnyak, A. V. et al. Augmentor α and β (FAM150) are ligands of the receptor tyrosine kinases ALK and LTK: hierarchy and specificity of ligand–receptor interactions. Proc. Natl Acad. Sci. USA 112, 15862–15867 (2015).
Mo, E. S., Cheng, Q., Reshetnyak, A. V., Schlessinger, J. & Nicoli, S. Alk and Ltk ligands are essential for iridophore development in zebrafish mediated by the receptor tyrosine kinase Ltk. Proc. Natl Acad. Sci. USA 114, 12027–12032 (2017).
Fadeev, A. et al. ALKALs are in vivo ligands for ALK family receptor tyrosine kinases in the neural crest and derived cells. Proc. Natl Acad. Sci. USA 115, E630–E638 (2018).
Reshetnyak, A. V. et al. Identification of a biologically active fragment of ALK and LTK-ligand 2 (augmentor-α). Proc. Natl Acad. Sci. USA 115, 8340–8345 (2018).
Crick, F. H. & Rich, A. Structure of polyglycine II. Nature 176, 780–781 (1955).
Warkentin, E. et al. A rare polyglycine type II-like helix motif in naturally occurring proteins. Proteins 85, 2017–2023 (2017).
Loren, C. E. et al. A crucial role for the Anaplastic lymphoma kinase receptor tyrosine kinase in gut development in Drosophila melanogaster. EMBO Rep. 4, 781–786 (2003).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Endres, N. F. et al. Conformational coupling across the plasma membrane in activation of the EGF receptor. Cell 152, 543–556 (2013).
Myers, K. V., Amend, S. R. & Pienta, K. J. Targeting Tyro3, Axl and MerTK (TAM receptors): implications for macrophages in the tumor microenvironment. Mol. Cancer 18, 94 (2019).
Grasberger, B., Minton, A. P., DeLisi, C. & Metzger, H. Interaction between proteins localized in membranes. Proc. Natl Acad. Sci. USA 83, 6258–6262 (1986).
Klein, P., Mattoon, D., Lemmon, M. A. & Schlessinger, J. A structure-based model for ligand binding and dimerization of EGF receptors. Proc. Natl Acad. Sci. USA 101, 929–934 (2004).
Kuriyan, J. & Eisenberg, D. The origin of protein interactions and allostery in colocalization. Nature 450, 983–990 (2007).
Diwanji, D., Thaker, T. & Jura, N. More than the sum of the parts: toward full-length receptor tyrosine kinase structures. IUBMB Life 71, 706–720 (2019).
Murray, P. B. et al. Heparin is an activating ligand of the orphan receptor tyrosine kinase ALK. Sci. Signal. 8, ra6 (2015).
Li, T. et al. Structural basis for ligand reception by anaplastic lymphoma kinase. Nature https://doi.org/10.1038/s41586-021-04141-7 (2021).
Van Duyne, G. D., Standaert, R. F., Karplus, P. A., Schreiber, S. L. & Clardy, J. Atomic structures of the human immunophilin FKBP-12 complexes with FK506 and rapamycin. J. Mol. Biol. 229, 105–124 (1993).
Monneau, Y. R. et al. Exploiting E. coli auxotrophs for leucine, valine, and threonine specific methyl labeling of large proteins for NMR applications. J. Biomol. NMR 65, 99–108 (2016).
Rossi, P., Monneau, Y. R., Xia, Y., Ishida, Y. & Kalodimos, C. G. Toolkit for NMR studies of methyl-labeled proteins. Methods Enzymol. 614, 107–142 (2019).
Xie, T., Saleh, T., Rossi, P. & Kalodimos, C. G. Conformational states dynamically populated by a kinase determine its function. Science 370, eabc2754 (2020).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Hendrickson, W. A. & Ogata, C. M. Phase determination from multiwavelength anomalous diffraction measurements. Methods Enzymol. 276, 494–523 (1997).
Terwilliger, T. C. et al. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr. D 65, 582–601 (2009).
Liebschner, D. et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D 75, 861–877 (2019).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. D 66, 486–501 (2010).
Delaglio, F. et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 (1995).
Lee, W., Tonelli, M. & Markley, J. L. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics 31, 1325–1327 (2015).
Lange, O. F. et al. Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples. Proc. Natl Acad. Sci. USA 109, 10873–10878 (2012).
Shen, Y. & Bax, A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J. Biomol. NMR 56, 227–241 (2013).
Rossi, P., Xia, Y., Khanra, N., Veglia, G. & Kalodimos, C. G. 15N and 13C- SOFAST-HMQC editing enhances 3D-NOESY sensitivity in highly deuterated, selectively [1H,13C]-labeled proteins. J. Biomol. NMR 66, 259–271 (2016).
Monneau, Y. R. et al. Automatic methyl assignment in large proteins by the MAGIC algorithm. J. Biomol. NMR 69, 215–227 (2017).
Guntert, P. Automated NMR structure calculation with CYANA. Methods Mol. Biol. 278, 353–378 (2004).
Linge, J. P., Williams, M. A., Spronk, C. A., Bonvin, A. M. & Nilges, M. Refinement of protein structures in explicit solvent. Proteins 50, 496–506 (2003).
Brunger, A. T. Version 1.2 of the Crystallography and NMR system. Nat. Protoc. 2, 2728–2733 (2007).
Bhattacharya, A., Tejero, R. & Montelione, G. T. Evaluating protein structures determined by structural genomics consortia. Proteins 66, 778–795 (2007).
Tejero, R., Snyder, D., Mao, B., Aramini, J. M. & Montelione, G. T. PDBStat: a universal restraint converter and restraint analysis software package for protein NMR. J. Biomol. NMR 56, 337–351 (2013).
Mastronarde, D. N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 152, 36–51 (2005).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Zivanov, J. et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018).
Grant, T., Rohou, A. & Grigorieff, N. cisTEM, user-friendly software for single-particle image processing. eLife 7, e35383 (2018).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Pettersen, E. F. et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Afonine, P. V. et al. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr. D 74, 531–544 (2018).
Williams, C. J. et al. MolProbity: more and better reference data for improved all-atom structure validation. Protein Sci. 27, 293–315 (2018).
Kortt, A. A., Nice, E. & Gruen, L. C. Analysis of the binding of the Fab fragment of monoclonal antibody NC10 to influenza virus N9 neuraminidase from tern and whale using the BIAcore biosensor: effect of immobilization level and flow rate on kinetic analysis. Anal. Biochem. 273, 133–141 (1999).
Zhao, H., Brautigam, C. A., Ghirlando, R. & Schuck, P. Overview of current methods in sedimentation velocity and sedimentation equilibrium analytical ultracentrifugation. Curr. Protoc. Protein Sci.71, 20.12.1–20.12.49 (2013).
Schuck, P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys. J. 78, 1606–1619 (2000).
Zhao, H. et al. A multilaboratory comparison of calibration accuracy and the performance of external references in analytical ultracentrifugation. PLoS ONE 10, e0126420 (2015).
Brautigam, C. A. Calculations and publication-quality illustrations for analytical ultracentrifugation data. Methods Enzymol. 562, 109–133 (2015).
Folta-Stogniew, E. & Williams, K. R. Determination of molecular masses of proteins in solution: implementation of an HPLC size exclusion chromatography and laser light scattering service in a core laboratory. J. Biomol. Tech. 10, 51–63 (1999).
Acknowledgements
This work was supported by the American Lebanese Syrian Associated Charities (ALSAC) and NIH grant R35 GM122462. Cryo-EM and NMR data were acquired at the St. Jude Cryo-EM Center and Biomolecular NMR Spectroscopy Center, respectively. X-ray data were collected at SERCAT ID and BM beamlines. We thank M. Rana and R. Kalathur for their help with protein expression and purification; M. Clay for assisting with NMR graphics; I. Chen for critical reading of the manuscript; E. Folta-Stogniew for discussions of SEC–MALS data; and Z. Luo for assisting with the preparation of figures and the animation.
Author information
Authors and Affiliations
Contributions
A.V.R. and C.G.K. supervised the project and wrote the manuscript. P.R., J.S. and A.G.M. contributed to the final version of the manuscript. A.V.R. and M.S. purified and prepared protein samples for NMR, X-ray, cryo-EM and biophysical studies. P.R. and M.S. performed NMR data collection, processing and structure calculations. A.V.R. and D.J.M. performed data collection and processing and solved the X-ray structures. A.V.R. and A.G.M. screened cryo-EM grids and performed cryo-EM data collection and processing. A.V.R. built atomic models for X-ray and cryo-EM studies and performed SEC–MALS experiments. A.V.R. and A.N. performed SV–AUC experiments and analysed data. I.L. and J.M. performed ALK cell-based phosphorylation assays. C.G.K. acquired funding and resources for the study.
Corresponding authors
Ethics declarations
Competing interests
C.G.K. is a consultant for BridgeBio.
Additional information
Peer review information Nature thanks the anonymous reviewers for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data figures and tables
Extended Data Fig. 1 Structural features of ALK-ECRABR.
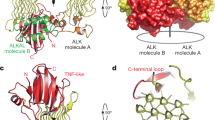
a, 1H-15N–correlated (left panel) and 1H-13C–correlated (right panel) spectra of [U-2H,15N; Ala-13CH3; Met-13CH3; Ile-δ1-13CH3; Leu,Val-13CH3/13CH3; Thr-13CH3]-labelled ALK-ECRABR. b, 1H-13C–correlated spectra of [U-2H; Phe-δ-13CH; Tyr-ε-13CH]-labelled ALK-ECRABR. c, Select strips from 13C-edited NOESY experiments highlighting intra-domain NOEs between TNF-like and EGF-like. d, Close-up view of the TNF-like−EGF-like interface and close-range inter-proton contacts (within ~7 Å) observed from the NOESY NMR analysis of spectra shown in (c). e, Asymmetric unit content of ALK-ECRABR(ΔEGF) crystals. f, 2Fo-Fc map of ALK-ECRABR(ΔEGF) depicted at 0.951 contour level for chain A. g, B-factors of ALK-ECRABR(ΔEGF) (chain A) are mapped on its structure. The tube radius is proportional to the B-factor. Low-to-high B-factors are also denoted in a blue-to-red colour gradient. h, Topology diagram of ALK-ECRABR(ΔEGF). PGII helices are shown in green tubes, β-strands in arrows and α helices in cylinders. i, Superposition of the following structures: ALK-ECRABR GlyR (grey, this work), glycine rich domain of GTP-binding protein Obg (red, PDB ID 1LNZ), acetophenone carboxylase (blue, PDB ID 5L9W), antifreeze protein (yellow, PDB ID 3BOG), and gp38 bacteriophage adhesin tip (green, PDB ID 6F45). j, Schematic representation of the GlyR PGII array shown in a top view. Solid hexagons denote PGII helices with an N-to-C direction towards the reader whereas open hexagons denote PGII helices in the opposite direction. k, Side view of the GlyR PGII array in ball-and-stick representation. l, Top view of GlyR PGII array showing inter-chain hydrogen bond network (grey dashes). m, Close-up view of the TNF-like−EGF-like interface.
Extended Data Fig. 2 ALK and LTK sequence comparison and structural characterization of ALKAL2.
a, Schematic representation of domain organization for human ALK and LTK receptors (left panel). Sequence alignment of ALK and LTK ECRs (right panel). The secondary structure diagram is shown based on the ALK-ECRABR structure determined in this work. Cys residues are coloured red and disulfide bridges are shown with red lines. Residues participating in ALKAL2 binding are coloured magenta, and residues participating in inter-protomer dimerization are underlined and coloured blue (contacts with TNF-like) and grey (contacts with THB). The key residues involved in ALKAL2 binding are conserved between ALK and LTK (highlighted with magenta in LTK sequence), with the exception of F143, S260, L361, Q362, A365, T367, E374, R376, D388, Q390 and L401 (LTK numbering, underlined with black lines). The difference in ALKAL2/1 specificity might be explained by the H120/Y99 and D124/E103 substitutions (ALKAL2/1 respectively) and/or difference in the receptor-receptor dimerization interface. b, SEC–MALS (upper panel) analysis of ALKAL2-AD at eluted concentrations of ∼67 μM (red), 11 μM (blue) and 1 μM (magenta). Molecular masses in kDa determined by in-line MALS (left axis) are included. SV–AUC profile of ALKAL2-AD (lower panel). Concentrations used are: 233.53 (purple), 111.25 (blue) and 55.62 µM (cyan). c, 1H-15N–correlated (left panel) and 1H-13C–correlated (right panel) NMR spectra of ALKAL2-AD. d, NMR ensemble of the 20 lowest-energy conformers of ALKAL2-AD. e, Electrostatic surface representation of ALKAL2-AD. The electrostatic potential is measured in eV, with range as shown in the corresponding colour bar (from −5.000 to +5.000 eV). f, NMR ensemble of the 20 lowest-energy conformers of ALKAL1-AD. g, SV–AUC profile and sedimentation coefficient distribution model c(s) of MBP–ALKAL2(C66Y) (left panel). Concentrations used are: 96.9 µM (purple), 48.5 µM (blue), 29.4 µM (cyan), 12.1 µM (green), 6.9 µM (yellow), 3.5 µM (orange) and 1.7 µM (red). Isotherm of the signal-weight-average s-values (sw) for MBP–ALKAL2(C66Y) obtained by integration of c(s) distributions over the s-range of 2.5 and 5 S for each loading concentration in a dilution series (right panel). The confidence intervals of the fits are presented in the lower panel. h, Superposition of NMR-solved and AlphaFold-predicted structures of ALKAL2 (left panel) and ALKAL1 (right panel). AD and variable region (VR) regions are labelled. i, SV–AUC profile of ALK-ECRABR–ALKAL2-AD. Concentrations used are: 177.8 (purple), 87.87 (blue), 43.93 (cyan), 20.92 (green), 10.46 (yellow), 5.44 (orange) and 2.72 µM (red). j, SV–AUC profile and sedimentation coefficient distribution model c(s) of MBP–ALKAL1. The highest (96.6 µM - purple) and lowest (4.8 µM - orange) concentrations are shown. k, SEC–MALS profile for the ALK-ECRABR−MBP–ALKAL1 complex (blue, theoretical mass of 91 kDa for 1:1 complex). The profile for the ALK-ECRABR−MBP–ALKAL2(C66Y) complex (red, theoretical mass of 189 kDa for 2:2 complex) is included for direct comparison. l, Sedimentation velocity analytical ultracentrifugation profile of ALK-ECRABR–MBP–ALKAL1 (loading concentration 74 µM).
Extended Data Fig. 3 Characterization of the binding mode and oligomeric state of the ALK-ECRABR–ALKAL2 complex.
a, SEC–MALS profiles of the ALK-ECRABR–ALKAL2(C66Y) complex. The corresponding isotherm of the signal-weight-average MW as a function of concentration is shown in the right panel. Standard errors were determined according to a previous study61. b, Sedimentation velocity analytical ultracentrifugation isotherm of the signal-weight-average s-values for ALK-ECRABR–ALKAL2(C66Y) complex. c, SEC–MALS profile (red) of ALK-ECRABR–MBP–ALKAL2(C66Y) complex mixed at 2:1 ratio. SDS/PAGE for corresponding fractions are shown in the right panel. Bands corresponding to ALK-ECRABR or MBP–ALKAL2(C66Y) are labelled, position of molecular weight markers are indicated. d, e, ALK autophosphorylation assays. d, ALK variants (as indicated in the labels) stably expressed in NIH/3T3 cells were stimulated with 10 nM of purified WT ALKAL2-AD. e, Wild type ALK stably expressed in NIH/3T3 cells was stimulated with 10 nM of purified ALKAL2-AD variants. Cell lysates were subjected to immunoprecipitation using anti-ALK antibodies followed by SDS/PAGE and immunoblotting with anti-pTyr (pY) and anti-ALK (ALK) antibodies. Relative position of the band for 180-kDa Mw marker is shown. f, SEC–MALS profiles of ALK-ECRABR(4M):ALKAL2(2M) complex. ALK-ECRABR(4M) stands for T686A/N787A/Q788A/I795A mutations in ALK-ECRABR, and ALKAL2(2M) for full-size ALKAL2(C66Y) containing I127A/Y130A mutations. The corresponding isotherm of the signal-weight-average MW as a function of concentration is shown in the right panel. Standard errors were determined according to61. g, Sedimentation velocity analytical ultracentrifugation isotherm of the signal-weight-average s-values for ALK-ECRABR(4M)–ALKAL(2M) complex.
Extended Data Fig. 4 Cryo-EM data processing workflow of ALK-ECRABR–ALKAL2(C66Y) and evaluation of the reconstruction.
Stages of cryo-EM data preprocessing, image classification and refinement for ALK-ECRABR–ALKAL2(C66Y) complex. Local resolution maps and fourier shell correlation are presented for final cryo-EM structures.
Extended Data Fig. 5 ALK-ECRABR rearrangements after ALKAL2 binding.
a, d, e, Superposition of ALKAL2 (a), GlyR-TNF-like (d) and EGF-like (e) structures in unliganded (pink) and liganded (blue) states. b, ALKAL2-induced repositioning of ALK-ECRABR. c, Cartoon representation of the heterotetrameric ALK-ECRABR–ALKAL2 complex wherein the 13-residue-long linker tethering EGF-like to TMH has been modelled in an extended conformation. The modelling shows that if EGF-like did not change its position upon ALKAL2 binding (EGF-like unliganded position shown in dark red) TMH dimerization would not be possible because the linker is too short. The model for the linker was manually built in Coot and follows the shortest possible path to reach the TMH. f, Superposition of EGF-like structures in the unliganded (orange) and ALKAL2-bound (grey) states demonstrates the conformational changes in EGF-like between the two states. g, Residues at the interface between EGF-like in unliganded state and TNF-like (left panel) and between EGF-like in ALKAL2-bound state and ALKAL2 (right panel). i, Wild type trx–ALKAL2-AD (grey) and trx–ALKAL2-AD variants (magenta) were tested for their ability to bind wild type ALK-ECRABR using BLI. Steady-state dissociation constants and standard errors were determined according to a previous study56. j, Comparison of tyrosine autophosphorylation of WT ALK stimulated by 10 nM of purified ALKAL2 variants as indicated. ALKAL2-ADRC stands for mutation of four charged residues - K94E/K96E/K99E/H100E; Trx–ALKAL2-AD is N-terminal fusion of ALKAL2 with thioredoxin, and ALKAL2-ADΔɑ1 is deletion of ɑ1 helix (residue boundaries 103-152). NIH3T3 cells stably expressing WT ALK were lysed after ALKAL2 stimulation and were subjected to immunoprecipitation using anti-ALK antibodies followed by SDS/PAGE and immunoblotting with anti-pTyr (pY) and anti-ALK (ALK) antibodies. Relative position of the band for 180-kDa Mw marker is shown. h, Close-up views of residues (CA atoms shown) D732, H996 and T733, M997 is shown. These residues were mutated to Cys for the cross-linking experiments.
Extended Data Fig. 6 NMR analysis of ALK-ECRABR–ALKAL2-AD and ALK-ECRABR–ALKAL1-AD complexes.
a, b, Superimposed 1H-13C–correlated spectra of ALK-ECRABR–ALKAL2-AD (a) and ALK-ECRABR–ALKAL1-AD (b) complexes. ALK-ECRABR and ALKAL proteins are 1H-13C labelled in the indicated methyl groups. c, d, Chemical shift perturbation induced by ALKAL1-AD binding to ALK-ECRABR to combined 1H and 15N amide atoms (c) and 1H and 13C methyl atoms (d). e, f, Chemical shift perturbation induced by ALKAL1-AD mapped onto the ALK-ECRABR structure. g, NMR characterization of ALKAL1-AD binding to ALK-ECRABR. Select strips from 13C-edited NOESY experiments showing intermolecular NOEs between ALK-ECRABR and ALKAL1-AD. Similar results were obtained when ALKAL2-AD was used, confirming that the structure observed in the frozen sample used in cryo-EM is the same in solution. h, NMR characterization of the EGF-like domain repositioning upon ALKAL1 binding to ECRABR. Select strips from 13C-edited NOESY experiments for ALK-ECRABR showing interdomain NOEs in the unbound form. Characteristic NOE patterns between Met997 of the EGF-like domain and the indicated residues of the TNF-like domain (right panel) changed dramatically upon ligand binding and demonstrate pronounced re-orientation of the EGF-like domain as shown schematically on the right panel.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1–3 and their accompanying legends.
41586_2021_4140_MOESM4_ESM.mp4
Supplementary Video 1 ALKAL-induced activation of ALK receptor. The video demonstrates how ALK is activated by monomeric ALKAL ligands (ALKAL1 or ALKAL1/2-AD).
Rights and permissions
About this article
Cite this article
Reshetnyak, A.V., Rossi, P., Myasnikov, A.G. et al. Mechanism for the activation of the anaplastic lymphoma kinase receptor. Nature 600, 153–157 (2021). https://doi.org/10.1038/s41586-021-04140-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41586-021-04140-8
This article is cited by
-
Multi-omic and functional analysis for classification and treatment of sarcomas with FUS-TFCP2 or EWSR1-TFCP2 fusions
Nature Communications (2024)
-
Autoinhibitory structure of preligand association state implicates a new strategy to attain effective DR5 receptor activation
Cell Research (2023)
-
A retrospective study of postoperative targeted therapy in ALK-positive lung cancer
Scientific Reports (2023)
-
Identification of Crucial Genes and Signaling Pathways in Alectinib-Resistant Lung Adenocarcinoma Using Bioinformatic Analysis
Molecular Biotechnology (2023)
-
Structural basis for ligand reception by anaplastic lymphoma kinase
Nature (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.