An in vivo Cell-Based Delivery Platform for Zinc Finger Artificial Transcription Factors in Pre-clinical Animal Models
- 1Department of Neurology, University of California Davis School of Medicine, Sacramento, CA, United States
- 2Stem Cell Program and Gene Therapy Center, University of California, Davis, Sacramento, CA, United States
- 3Department of Biochemistry and Molecular Medicine, Genome Center, University of California, Davis, Davis, CA, United States
- 4Department of Psychiatry and Behavioral Sciences, MIND Institute, University of California Davis School of Medicine, Sacramento, CA, United States
- 5Departments of Pediatrics and Cell Biology and Human Anatomy, School of Medicine, Gene Therapy Center, and California National Primate Research Center, University of California, Davis, Davis, CA, United States
- 6Department of Otolaryngology, University of California, Davis, Davis, CA, United States
Zinc finger (ZF), transcription activator-like effectors (TALE), and CRISPR/Cas9 therapies to regulate gene expression are becoming viable strategies to treat genetic disorders, although effective in vivo delivery systems for these proteins remain a major translational hurdle. We describe the use of a mesenchymal stem/stromal cell (MSC)-based delivery system for the secretion of a ZF protein (ZF-MSC) in transgenic mouse models and young rhesus monkeys. Secreted ZF protein from mouse ZF-MSC was detectable within the hippocampus 1 week following intracranial or cisterna magna (CM) injection. Secreted ZF activated the imprinted paternal Ube3a in a transgenic reporter mouse and ameliorated motor deficits in a Ube3a deletion Angelman Syndrome (AS) mouse. Intrathecally administered autologous rhesus MSCs were well-tolerated for 3 weeks following administration and secreted ZF protein was detectable within the cerebrospinal fluid (CSF), midbrain, and spinal cord. This approach is less invasive when compared to direct intracranial injection which requires a surgical procedure.
Introduction
Historically, engineered stem cell therapies have been utilized as a gene replacement and cross-correction approach by producing proteins at supraphysiologic levels in vivo (Kohn et al., 1995, 1998, 2021; Visigalli et al., 2010; Pollock et al., 2016; Adhikari et al., 2019, 2021; Beegle et al., 2020). Mesenchymal stem/stromal cells (MSCs) are a particularly favorable cell type as they are easily obtained, expanded, and can be readily manipulated ex vivo (Meyerrose et al., 2010; Damasceno et al., 2020). Additionally, MSCs do not permanently engraft into the host tissue (Kean et al., 2013), do not induce immune responses through the secretion of immunomodulatory cytokines, and demonstrate a clinically favorable safety profile (Thompson et al., 2020). These unique characteristics of MSCs make them an attractive potential delivery vehicle for gene modifying artificial transcription factors (ATF) such as zinc finger (ZF), transcription activator-like effectors (TALE), and CRISPR/Cas9 (Russ and Lampel, 2005; Thakore et al., 2015; Bailus et al., 2016; Fink et al., 2016; Halmai et al., 2020). The identification of an effective delivery system remains a challenge in the gene therapy field as these gene modifying approaches become increasingly sophisticated for the treatment of genetic disorders. Direct administration of purified protein (Bailus et al., 2016; Perdigão et al., 2020), lipid nanoparticle (Wang et al., 2016; Cheng et al., 2020), and viral-mediated approaches such as adeno-associated virus (Mendell et al., 2017; Russell et al., 2017) have been explored as putative delivery systems for ATF in vivo; however, a cell-based system such as MSCs has not been reported to date.
The most common cause for Angelman Syndrome (AS) is the loss of UBE3A in mature neurons due to a deletion of the maternal UBE3A allele and silencing of the paternal allele via a long non-coding RNA (Kishino et al., 1997; Chamberlain and Lalande, 2010). We previously described the use of a systemically administered, purified ZF-KRAB protein that achieved brain-wide distribution and activation of paternal Ube3a in a mouse model of AS by targeting ZF to the Snurf/Snrpn locus, a putative promoter for the Ube3a- antisense transcript (Landers et al., 2004; Bailus et al., 2016). Here, we evaluate engineered MSCs as a novel delivery platform for ZF gene modifiers into the central nervous system (CNS) of pre-clinical mouse models and juvenile rhesus monkeys. We observed that ZF secreting MSCs can secrete full-length, biologically active protein in vitro. Secreted ZF functionality was evaluated in two mouse models of AS: a paternal Ube3a reporter mouse (Ube3a+/yfp) and a maternal Ube3a deletion model (Ube3amat–/pat+). Both intracranial and cisterna magna (CM) injection of ZF-MSC resulted in protein uptake in neuronal tissue and activation of a paternal Ube3a reporter across multiple brain regions in mice 3 weeks following injection. ZF-MSC treatment in the Ube3amat–/pat+ AS mouse model also resulted in attenuated motor deficits up to 29 days post-injection. Lastly, studies were conducted in young rhesus monkeys to evaluate the translational feasibility, safety, and tolerability of ZF-MSCs in a pre-clinical animal model with close similarities to human anatomy and physiology. ZF-MSCs were well tolerated following intrathecal (IT) administration. Secreted ZF protein was detected in the cerebrospinal fluid (CSF), midbrain, and spinal cord 3 weeks following administration of ZF-MSCs. Overall, this study demonstrates the functionality of an MSC-based secretion platform for biologically active ATF in vivo.
Materials and Methods
Vector Design and Lentiviral Packaging
IgK-Leader sequence with TATp was Gibson cloned into the original pMAL-TatP-mCherry-HA-NLS-ZF00-KRAB via SalI and AscI. The transgene was excised from pMAL backbone via SalI (NEB, Ipswich, MA, United States) and PstI (NEB), followed by blunting with DNA Polymerase I (NEB) cloned into our pCCLc-MNDU3-X2-WPRE vector via SmaI (NEB) restriction site. Clones were subsequently Sanger sequenced confirmed (Quintara Biosciences, South San Francisco, CA, United States). Lentivirus was prepared accordingly to previously published protocols (Kalomoiris et al., 2012). Briefly, lentivirus was generated by complexing 25 μg of transgene vector, 25 μg of delta 8.9, and 5 μg were VSVG with 145 μg of PEI in 3 ml serum-free DMEM-HG (Thermo Fisher Scientific, Waltham, MA, United States, Catalog #11965118) for 20 min prior to application to 2.5 × 106 Lenti-X 293 cells (Takara Bio, Kusatsu, Japan, Catalog #632180). Twenty-four hours following transfection, cells were switched to serum-free Ultraculture (Lonza, Basel, Switzerland, Catalog #BP12-725F) for 48 h. Viral harvest was performed by transferring the supernatant of transfected Lenti-X 293 cells into 50 ml conical tubes, centrifuged at 500 RPM for 5 min to pellet cellular debris, and virus was isolated in a 100 kDa Centricon (MilliporeSigma, Burlington, MA, United States, Catalog #UFC710008).
Cell Isolation and Culturing
Mouse Bone-Marrow Mesenchymal Stem/Stromal Cell
The extraction protocol for obtaining MSCs was adapted from previously published procedures (Rossignol et al., 2011). Briefly, bone marrow was aspirated from the fibula of adult (6–8-week-old) C57BL/6 mice (Jackson Laboratory, Bar Harbor, ME, United States, Catalog # C57BL/6J) using a 25-gauge needle and attached 3 ml syringe. The cells were then suspended in 10 ml MSC medium (alpha modified Eagle’s medium, Sigma, St Louis, MO, United States, Catalog # D5030) with 20% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA, United States, Catalog #16000044) and 5 mg/ml streptomycin and 5 UI/ml penicillin (Sigma, Catalog #85886). Following incubation for 48 h at 37°C, 5% CO2, and 20% O2 the medium was replaced with fresh MSC medium to remove non-adherent cells. MSCs were passaged, expanded, and cryopreserved over passages 3–8. Passage 5 MSCs were transduced with competent lentivirus at MOI 100 and confirmed for transduction via fluorescent microscopy and sequencing. Vector copy number/cell was determined as WPRE/2GAPDH and verified as ∼2–5 vector transgene copies per cell. Primers/probes were designed based on published sequences and are as follows: moGAPDH- 5′-ACC ACG AGA AAT ATG ACA ACT-3′, 5′-CCC ACT GCC TAC ATA CCA TGA-3′, and 5′/56-FAM/TCA GCA ATG CAT CCT GCA CCA CC/36-TAMSp/3′. WPRE—5′-TTA CGC TAT GTG GAT ACG CTG-3′, 5′-TCA TAA AGA GAC AGC AAC CAG G-3′, and 5′/56-FAM/AGG AGA AAA TGA AAG CCA TAC GGG AAG C/36-TAMSp/3′. Quantitative PCR was performed using TaqMan Universal PCR Master Mix, no AmpErase UNG (Life Technologies, Catalog# 4304437), and the indicated primers/probes under the following reaction conditions on an Applied Biosystems StepOne Plus (Thermo Fisher Scientific, Waltham, MA, United States, Catalog #4376600): Stage 1—50°C for 2 min, Stage 2—95°C for 10 min, Stage 3—40 cycles of 95°C for 15 s and 60°C for 1 min, and Stage 4 (dissociation)—95°C for 15 s, 60°C for 1 min, 95°C for 15 s, and 60°C for 15 s. Quantification was based on standard curves of plasmid DNA.
Mesenchymal Stem/Stromal Cell Characterization
For characterization for MSC by flow cytometry, cells were lifted by TrypLE and resuspended in PBS. Cells were then incubated with the respective conjugated antibodies individually (all diluted 1:100 in PBS, Supplementary Table 1) for 30 min at 4°C. For CD31, cells were washed once with PBS and then stained for additional 15 min with an PE-conjugated anti-rat IgG antibody. Then, all samples were washed once with 1 ml of PBS, centrifuged for 300 × g for 5 min and resuspended in PBS prior to analysis using a Attune flow cytometer. Negative controls (conjugated isotype antibodies and unlabeled cells) were used for gating.
Secreted Zinc Finger Protein Confirmation
Transduced MSCs were washed twice with PBS and incubated with OptiMem (Thermo Fisher Scientific, Waltham, MA, United States, Catalog #31985070) for 48 h before being concentrated in a 10-kDa Amicon ® Ultra-15 Centrifugal Filter Unit (MilliporeSigma, Burlington, MA, United States, Catalog #UFC9030). This conditioned media was probed for presence of secreted protein with ms-HA (1:1,000, BioLegend, San Diego, CA, United States, Catalog #16B12) and Rb anti-β-Tubulin (1:2,000, Thermo Fisher Scientific, Waltham, MA, United States, Catalog #AA10).
Transwell Experiments
Neuro2As (ATCC, Manassas, VA, United States, Catalog #CCL-131) were cultured in DMEM-HG (Thermo Fisher Scientific, Waltham, MA, United States, Catalog #11965092), 10% FBS, and 1% L-Glutamine. 5 × 104 Neuro2As were seeded per well in 12-well Transwell plates (Corning, Corning, NY, United States, Catalog #3401) and 2.5 × 104 were seeded in Transwell baskets. Cells were equilibrated in their respective media for 24-h prior to co-incubation in reduced-serum Opti-Mem (Thermo Fisher Scientific, Waltham, MA, United States, Catalog #51985091) for longitudinal qPCR experiments. Purified ZF-P was provided by Dr. David Segal and spiked into Neuro2A cells at a dose of 100 μg/well. RNA was isolated using the Direct-Zol RNA Isolation Kit (Zymo Research, San Diego, CA, United States, Catalog #R2053). cDNA synthesis was performed using the RevertAid Random Hexamer Kit (Thermo Fisher Scientific, Waltham, MA, United States, Catalog #K1622). Ten nanograms of cDNA was probed with Snrpn primers (Snrpn-F 5′-TGCTACGTGGGGAGAACTTG-3′, Snrpn-R 5′-CCTGGGGAATAGGTACACCTG-3′) and Gapdh primers (Gapdh-F5′-TGACCACAGTCCATGCCATC-3′, Gapdh-R GACGGACACATTGGGGGTAG) with PowerUp SYBR Green (Thermo Fisher Scientific, Waltham, MA, United States, Catalog #A25741) in the Applied Biosystems StepOne Plus (Thermo Fisher Scientific, Waltham, MA, United States, Catalog #4376600). Data presented as 2–ΔΔCt were normalized to the NT-MSC control group.
Primary Cortical Neuron Isolation and Culture
Primary cell cultures of cortical neurons were prepared from E15.5 Ube3a+/yfp prenatal mice. Brains were dissected and cortices isolated in ice-cold PBS (100 units/mL penicillin-streptomycin). Cortical tissue was dissociated by enzymatic digestion [50 units papain (Worthington, Catalog # LK003172), 250 units DNase (Sigma, Catalog #D4513) 1 mM L-cysteine, 0.5 mM EDTA (Sigma, Catalog #P0899) in HBSS] and mechanical trituration. Cells were filtered by density gradient centrifugation [HBSS, ovomucoid protease inhibitor (Worthington, Catalog #KL003182) with bovine serum albumin, BSA], counted, and plated on poly-D-lysine (Sigma, Catalog #A3890401) coated 12-well plates at 1 × 106 cells per well. Cultures were grown in Neurobasal medium (Invitrogen, Catalog #21103-049) with 1× B-27 Supplement (Invitrogen) and 2 mM L-glutamine (Invitrogen, Catalog #25030-081) at 37°C and 5% CO2. At the first media change (72 h), 1 μM cytosine β-D-arabinofuranoside hydrochloride (Sigma, Catalog #C66415) was added to prevent replication of non-neuronal cells.
Immunocytochemistry
Mesenchymal Stem/Stromal Cell
Transduced MSCs were washed with PBS and fixed using 10% formalin fixation for 10 min on ice. MSCs were blocked in 10% SEABLOCK (Thermo Fisher Scientific, Catalog #37527) + PBS + 0.1% Triton X100 (VWR, Catalog #0694-1L) for 1 h prior to incubation with primary antibody 1:500 rb-mCherry (abcam, Boston, MA, United States, Catalog #ab167453) and 1:1,000 ms-Nestin (abcam, Catalog# 22035) for 1.5 h. MSCs were rinsed 3× with ice cold PBST and incubated with AlexaFluor secondary antibody 1:1,000 goat rb-594 (Thermo Fisher Scientific, Catalog #A32740) and 1:1,000 goat ms-488 (Thermo Fisher Scientific, Catalog #A28175) for 1.5 h and then with 1:1,000 Hoechst (Cell Signaling, Catalog #4082) for 10 min. MSCs were then rinsed 2× with PBST before storing in PBS. Cells were imaged on a Nikon Eclipse Ti-U inverted research microscope (Nikon, Melville, NY, United States).
Primary Cortical Neurons
Treated neurons were washed with PBS and fixed using 10% formalin fixation for 10 min on ice. Neurons were blocked in 10% SEABLOCK (Thermo Fisher Scientific, Catalog #37527) + PBS + 0.1% Triton X100 (VWR, Catalog #0694-1L) prior to incubation with primary antibody 1:500 rb-GFP (1:1,000, Novus Biologics, Centennial, CO, United States, Catalog #NB600-308) for 1 h. Neurons were rinsed 3× with ice cold PBST and incubated with secondary antibody 1:1,000 goat rb-488 (Alexa Fluor, Catalog #A32731) for 1 h. Neurons were then rinsed 2× with PBST and imaged on a Nikon Eclipse Ti-U inverted research microscope (Nikon, Melville, NY, United States).
Mice
All experimental procedures were performed in accordance with the National Institutes of Health Guide for Care and Use of Laboratory Animals and were approved by the Institutional Animal Care and Use Committees (IACUC) of the University of California, Davis. A total of 81 mice were used for the following studies. Eleven FVB mice were used for initial MSC secretion characterization studies (7F, 4M) (Figure 3). Three Ube3matYfp/pat+ were used as positive controls (2F, 1M) (Figures 3–5). Twenty-one Ube3a+/yfp mice were used for 1-week co-localization studies (4F, 5M) and 3-week molecular studies (6F, 6M) (Figures 3–5). Forty-six Ube3amat–/pat+ were used for behavioral studies (21F, 25M) (Figure 6). Experimenters were blinded to treatment groups and genotypes until final data analysis was performed. All studies were sex balanced when possible.
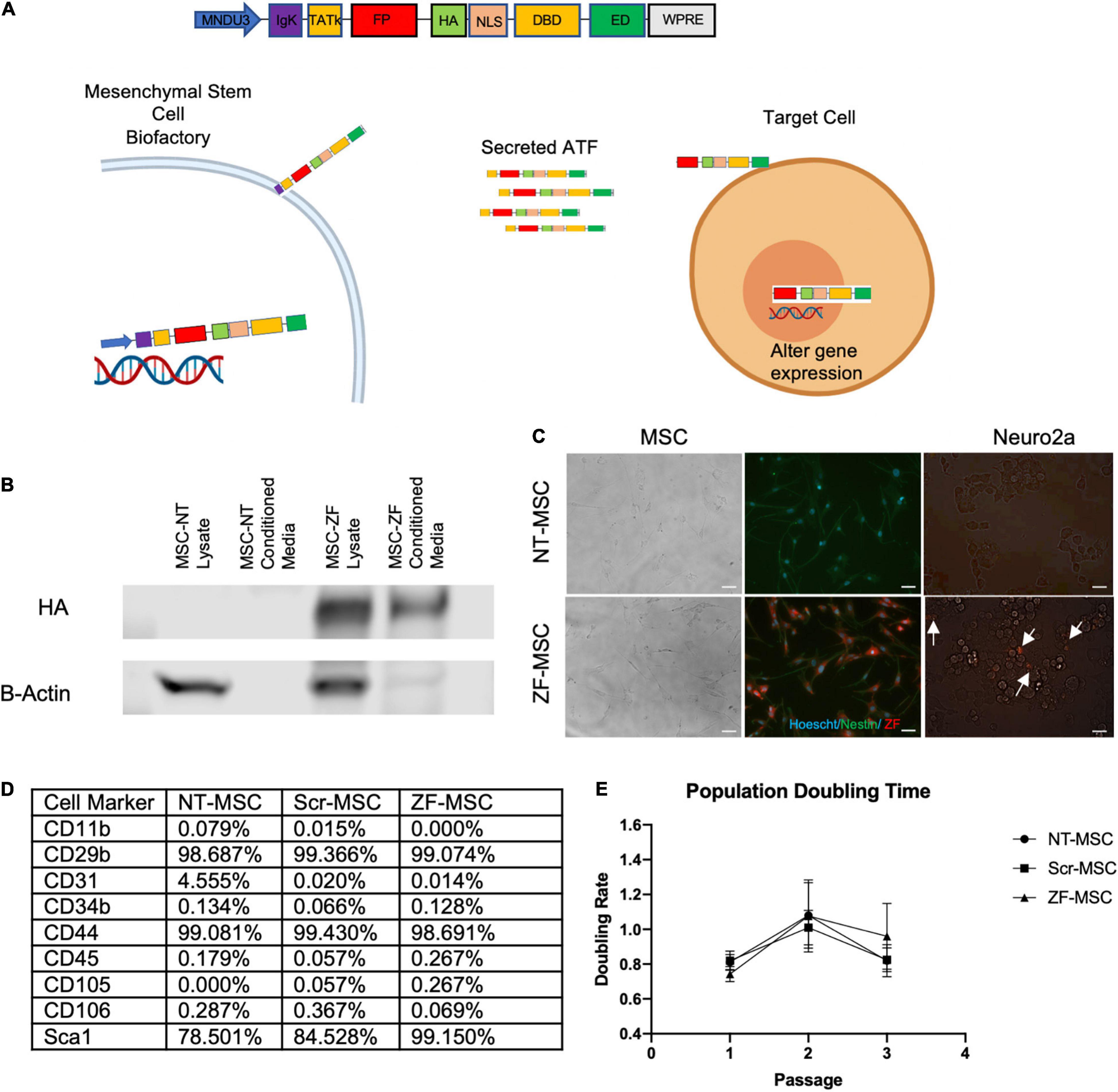
Figure 1. Engineered MSC can function as biofactories for zinc finger (ZF). (A) Schematic of design of secreted protein. Injected MSC biofactories secrete ZFs via the IgK Leader Signal Peptide into the extracellular space whereby these proteins are taken up by neighboring cells by the TATk element. ZFs are trafficked into the nucleus of target cells via NLS-tagged transport to alter target cell gene expression. (B) Western blot depicting presence of HA-tagged secreted ZF in conditioned media of MSC 48-h following serum starvation. (C) Left Panel: Fluorescent microscopy depicting Hoechst-labeled MSCs (blue) transduced to express ZF (red). Right Panel: Fluorescent microscopy depicting co-localization of ZF (red) on recipient Neuro2As following a 24-h incubation with conditioned media from ZF-MSCs. White arrows indicate Neuro2A that co-localize with ZF. Scale bar = 100 μm. (D) Table summary of FACS detailing the enrichment of isolated and transduced MSCs for typical MSC markers and negative for markers of cells of hemopoietic origin, related to Supplementary Figures 2, 3. (E) MSCs were passaged every 2 days and cell counts were recorded prior to plating. Transduction of MSCs to secrete either a Scr ZF or ZF did not demonstrate altered population doubling times compared to an isogenic NT-MSC. Error bars indicate ± standard error of the mean. Dots indicate average mean of four biological replicates. Two-Way ANOVA followed by a Tukey’s HSD. MNDU3, endogenous promoter; IgK, secretion peptide; TATk, furin-resistant TAT peptide; FPR, fluorescent protein reporter; HA, hemagglutinin epitope; NLS, nuclear localization signal; DBD, DNA-binding domain; ED, effector domain; WPRE, woodchuck hepatitis virus post-transcriptional element; ZF-MSC, MSCs that have been transduced to secrete ZF; NT-MSC, non-transduced MSC.
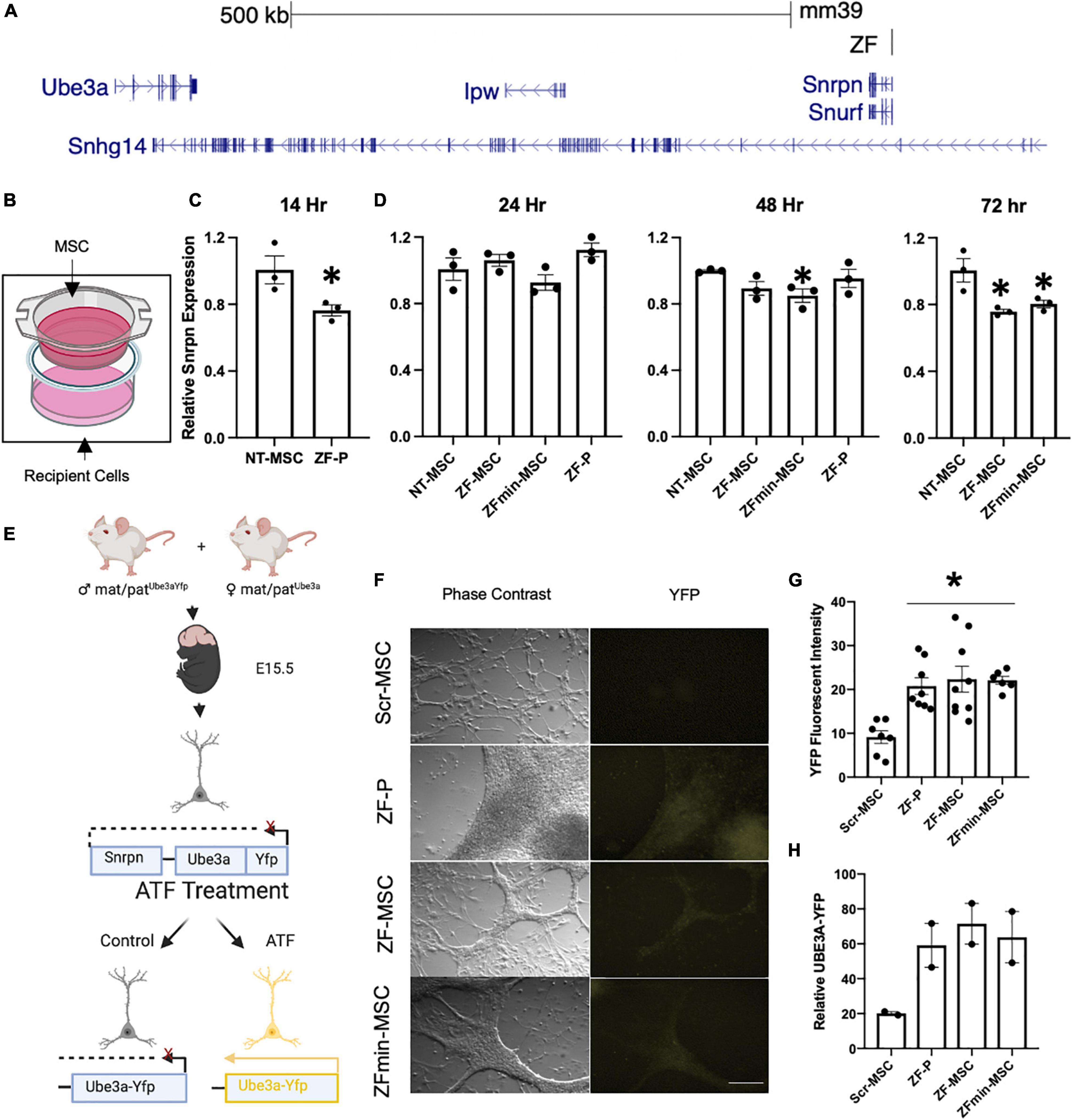
Figure 2. Secreted ZF protein demonstrates transcriptional effect in genes known in angelman syndrome. (A) Schematic of the ZF Binding to the putative promoter of the long non-coding RNA in the imprinted paternal Ube3a allele. (B) Schematic of transwell experiment, ZF-MSCs are seeded in the upper Transwell basket while recipient Neuro2As are seeded in the bottom well. (C,D) The efficacy of secreted ZF from MSC to repress Snrpn was assessed longitudinally in Neuro2a cells as measured by RT-qPCR. Each dot is representative of independent well. n = 3 independent wells per group. (C) An observable reduction of Snrpn expression was observed following a 14 h incubation with 100 μg purified ZF-P as a positive control for Snrpn, Student T-test, *p < 0.05 to NT-MSC repression. (D) An amelioration of ZF-P repression of Snrpn is observed by 24 h whereas ZF-MSC and ZFmin-MSC co-incubated groups demonstrate an observable decline of Snrpn expression over 48 h, [F(3, 8) = 2.738] *p < 0.05 to NT-MSC, to 72 h, [F(2, 6) = 9.225], *p < 0.05 to NT-MSC. (E) Schematic of experimental design for screening ZF in primary cortical neurons derived from E15.5 Ube3a+/yfp pups. Successful interference of long non-coding RNA transcription activates paternally silent Ube3ayfp. (F) Immunocytochemistry panel of representative Ube3a+/yfp primary cortical neurons shows activation of UBE3A-YFP (yellow) following incubation 24 h incubation with 250 μg of ZF-P, ZF-MSC CM, or ZFmin-MSCCM. Scale bar = 100 μm. (G) Quantification of YFP mean fluorescent intensity from H showed a significant two-fold increase in YFP fluorescent intensity in immune-labeled Ube3a+/yfp primary cortical neurons. One-Way ANOVA [F(3, 26) = 7.955] followed by Fisher’s LSD, *p < 0.05 from NT-MSC. Each dot is representative of an independent well containing an isolated cortex from a Ube3a+/yfp pup. n = 7–9 wells per group. (H) Observable increases of UBE3A-YFP were visualized in western blot analysis of treated primary cortical neurons. Each dot is representative of an independent well containing an isolated cortex from a Ube3a+/yfp pup. n = 2 wells per group. Yfp, yellow fluorescent protein. ZF-P, purified ZF protein. Error bars indicate ± standard error of the mean.
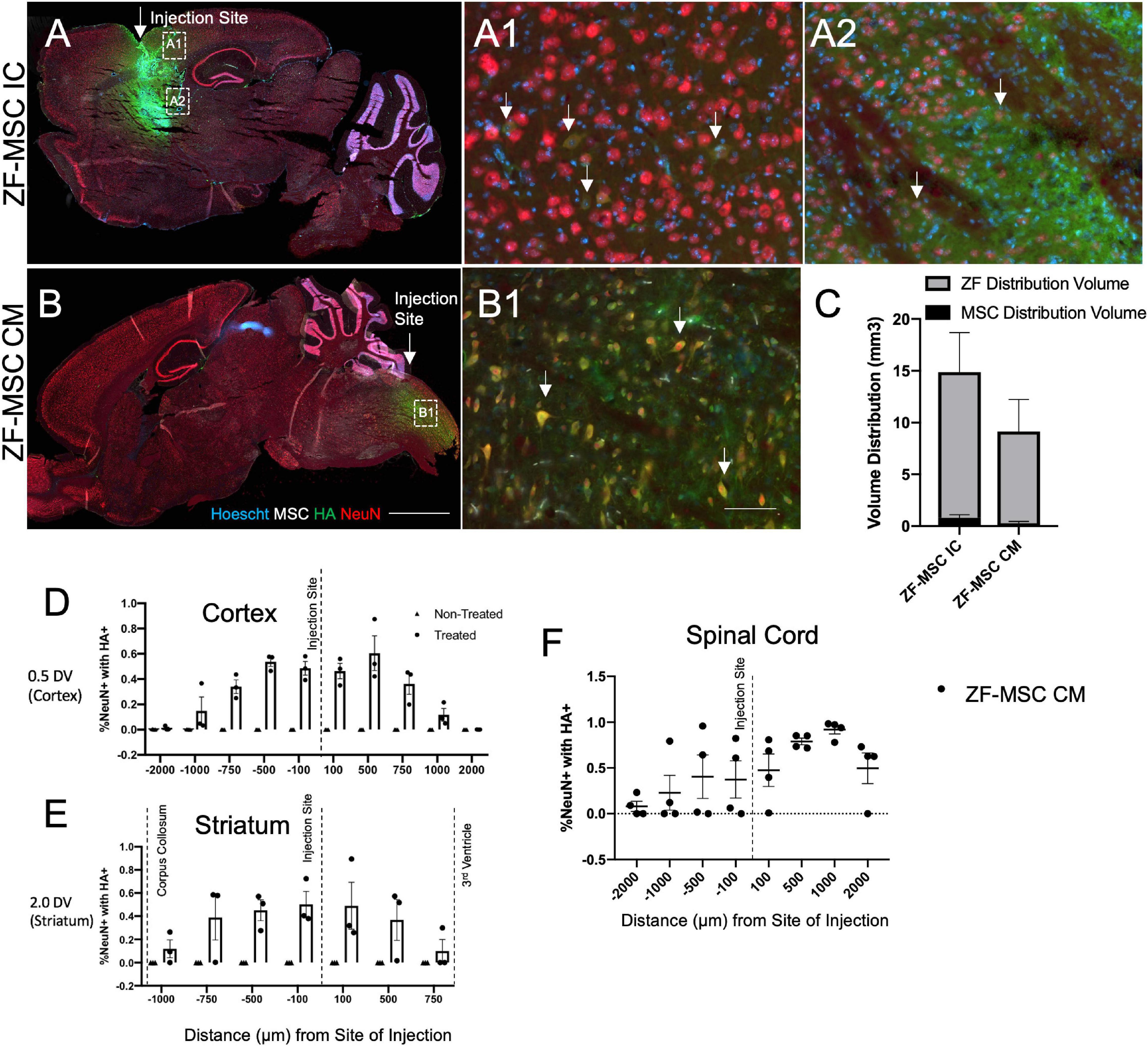
Figure 3. Secreted zinc finger (ZF) diffuse in a gradient fashion from site of injection. (A) Intracranial injection of ZF-MSC into the striatum resulted in an observable ZF (red) distribution. Uptake of ZF is observable in mature NeuN+ (red) neurons in the cortex (A1) and the striatum (A2). White arrows indicate ZF+/NeuN+ cells. (B) CM injection of ZF-MSC demonstrated observable ZF distribution and uptake of protein NeuN+ neurons in the spinal cord (B1). White arrows indicate ZF+/NeuN+ cells. (C) Quantitation of volume distribution of ZF+ protein in the brain from injected cells from panels (A,B). The gray bar indicates ZF distribution. The black bar indicates the volume of ZF-MSC bolus injection. (D) Quantitation of co-localization of ZF protein with NeuN+ cells. ZF/NeuN co-localization decreased from the site of injection in the cortex. (E) Quantitation of co-localization of ZF protein with NeuN+ cells. ZF/NeuN co-localization decreased from the site of injection in the striatum. Dashed lines indicate appearance of morphologically distinct neuronal landmarks. Error bars indicate ± standard error of the mean. (F) Quantitation of co-localization of ZF protein with NeuN+ cells. ZF/NeuN co-localization decreased in the spinal cord adjacent to the location of the CM administration. DV, dorsal ventral; NeuN+, neuronal nuclei; HA, hemagglutinin. Scale bar = 1,000 μm (A,B), 100 μm (A1,A2,B1).
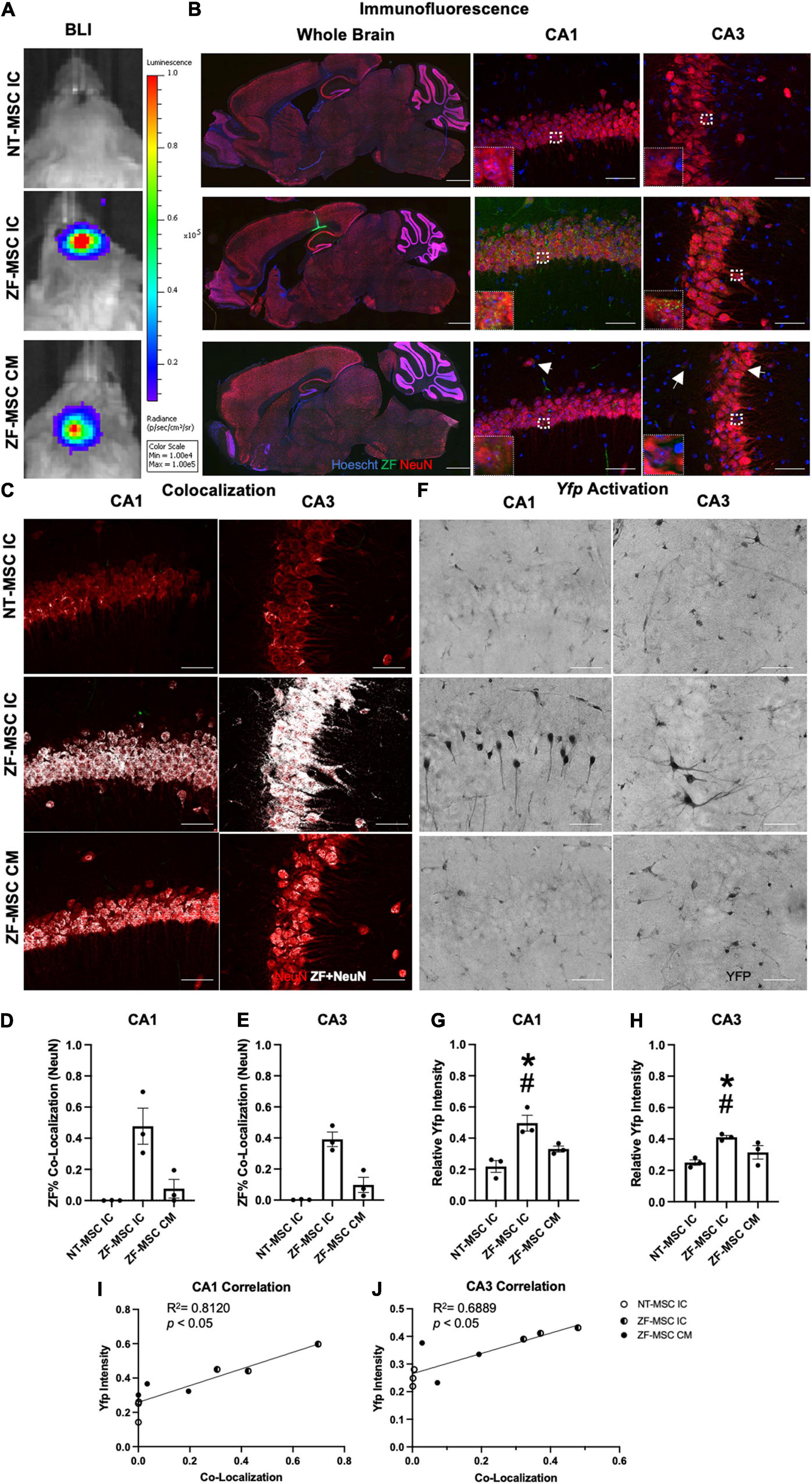
Figure 4. ZF-MSC persist 1 week following injection and demonstrate In vivo protein uptake. (A) Transduced ZF-MSC are detectable at the surgical site within the brain (ZF-MSC IC) or at the base of the skull (ZF-MSC CM) via bioluminescent imaging (BLI) 7 days following administration. (B) (Left Column) Bolus of ZF-MSCs (green) were observed within the corpus callosum, dorsal to the hippocampus of IC-injected mice 7 days following transplantation. Scale bar = 1,000 μm. Observable ZF+ (green) puncta are observed within the mature NeuN+ (red) neurons in CA1 (middle column) and CA3 (right column) of the hippocampus in both IC and CM-injected mice. White arrows indicate zinc finger (ZF). Scale bar = 50 μm. Inlet images are 20 μm × 20 μm of indicated region in parent image and depicts ZF+ puncta. (C–E) Co-localization of ZF+ (white) were observed in NeuN+ neurons in CA1 (D) and CA3 (E). Dots represent individual Ube3a+/yfp mice. n = 3 mice per group. (C) Representative images of CA1 (middle row) and CA3 (bottom row) depicting co-localization of ZF+ protein mature neurons. White dots indicate co-localized mCherry+ pixels with red NeuN pixels from images generated in Figure 2B. (D) ZF-MSC IC showed a 46.8% co-localization and ZF-MSC CM showed a 7.6% ZF co-localization with NeuN+ neurons across the entire CA1 region of the hippocampus. One-Way ANOVA [LSD F (2, 6) = 11.52] followed by a Fisher’s LSD, *p < 0.05 from NT-MSC. (E) ZF-MSC IC showed a 39.1% co-localization and ZF-MSC CM showed a 9.8% ZF co-localization with NeuN+ neurons across the entire CA3 region of the hippocampus. One-Way ANOVA [F(2, 6) = 26.82] followed by a Fisher’s LSD, *p < 0.05 from NT-MSC. (F–J) An increase in UBE3A-YFP was observed in CA1 (G) and CA3 (H) following IC administration of ZF-MSC. Graphical values normalized to a maternal Ube3aYfp positive control. Error bars indicate ± standard error of the mean. Dots represent individual Ube3a+/yfp mice. n = 3 mice per group. (F) Representative DAB-labeled images depicting morphologically distinct UBE3A-YFP+ neurons within CA1 (middle row) and CA3 (bottom row) of IC-administered ZF-MSC. Scale bar = 50 μm. (G) Quantitation of UBE3A-YFP mean fluorescence intensity of ZF-MSC IC and ZF-MSC CM administration in CA1 of Ube3a+/yfp mice 1 week following cell injection. ZF-MSC IC significantly increased UBE3A-YFP by 27.8% when compared to NT-MSC IC. One-Way ANOVA [F(2, 7) = 14.66] followed by Fisher’s LSD, *p < 0.05 from NT-MSC IC. #p < 0.05 from ZF-MSC CM. (H) Quantitation of UBE3A-YFP mean fluorescence intensity of ZF-MSC IC and ZF-MSC CM administration in CA3 of Ube3a+/yfp mice 1 week following cell injection. ZF-MSC IC significantly increased UBE3A-YFP by 16.1% when compared to NT-MSC IC. One-Way ANOVA [F(2, 7) = 5.747] followed by Fisher’s LSD, *p < 0.05 from NT-MSC IC. #p < 0.05 from ZF-MSC CM. (I) YFP expression is strongly and positively correlated with increased ZF co-localization in CA1 from (D,G). [F(1, 7) = 11.50] p = 0.009, R2 = 0.8120. (J) YFP expression is strongly and positively correlated with increased ZF co-localization in CA3 from (E,H). [F(1, 7) = 20.19] p = 0.0056, R2 = 0.6889.
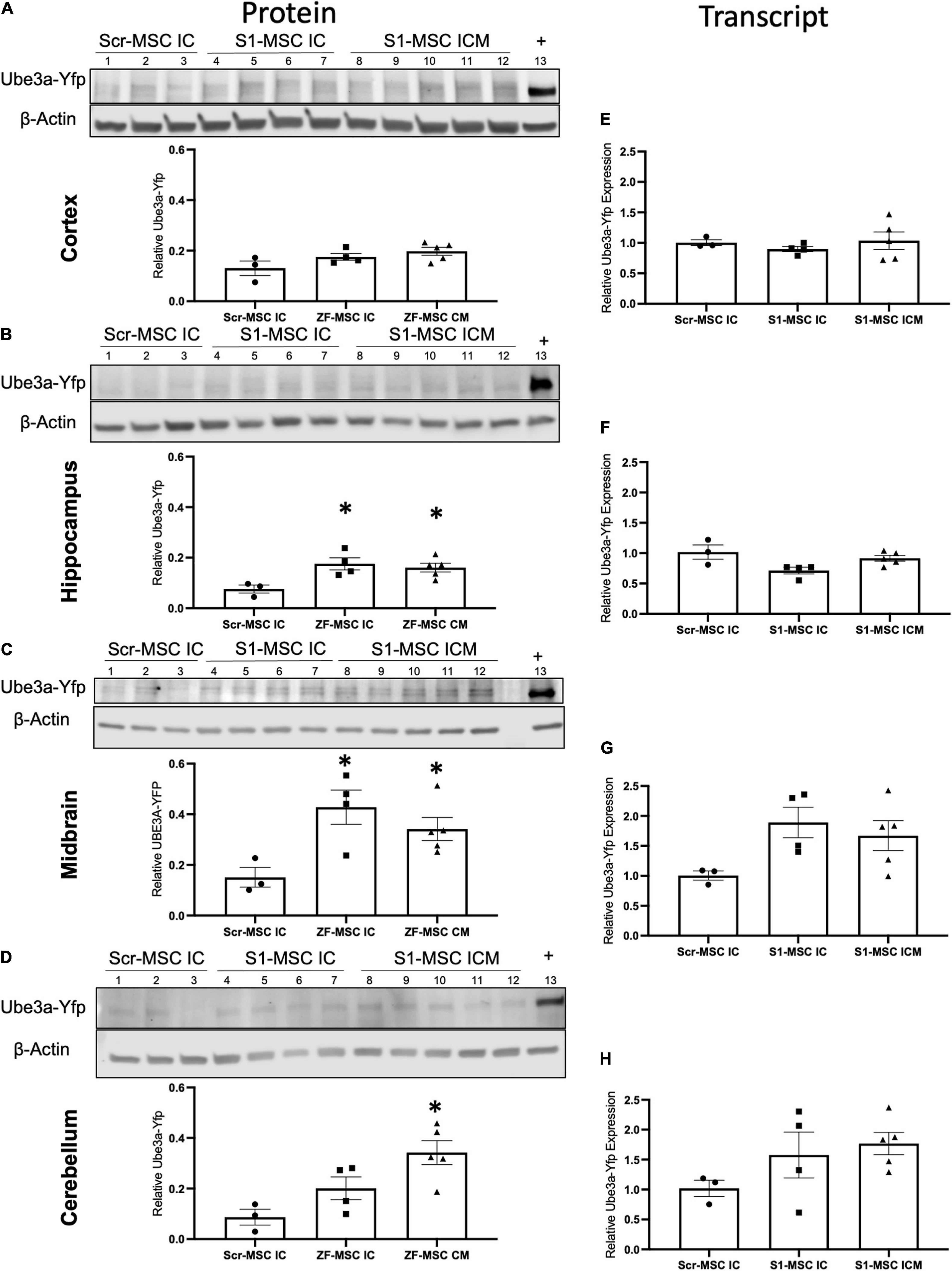
Figure 5. ZF-MSC demonstrate route of administration-dependent activation of Ube3aYfp 3 weeks following injection. (A–D) Representative Western blots of neuronal subregions with corresponding quantitative graphs. Graphical values normalized to a maternal Ube3aYfp positive control. Individual dots represent a single mouse. n = 3–5 mice per group. Lanes 1–3 = Scr-MSC IC. Lanes 4–7 = ZF-MSC IC. Lanes 8–12 = ZF-MSC ICM. Lane 13 = Positive YFP control. (A) No significant changes in cortical UBE3A-YFP protein expression were found following either ZF-MSC IC or ZF-MSC CM treatment. One-Way ANOVA [F(2, 9) = 3.103] followed by Fisher’s LSD, *p < 0.05 to NT-MSC. (B) A significant increase in hippocampal UBE3A-YFP protein expression was observed in both ZF-MSC IC and ZF-MSC CM treated mice. One-Way ANOVA [F(2, 9) = 6.041] followed by Fisher’s LSD, *p < 0.05 to NT-MSC. (C) A significant increase in midbrain UBE3A-YFP protein expression was observed in both ZF-MSC IC and ZF-MSC CM treated mice. One-Way ANOVA [F(2, 9) = 7.658] followed by Fisher’s LSD, *p < 0.05 to NT-MSC. (D) A significant increase in cerebellar UBE3A-YFP protein expression was observed with ZF-MSC CM treated mice. One-Way ANOVA [F(2, 9) = 5.717] followed by Fisher’s LSD, *p < 0.05 to NT-MSC. (E–G) Quantitative PCR of neuronal subregions. (E) No significant changes in cortical Ube3aYfp transcript expression were found following either ZF-MSC IC or ZF-MSC CM administration. One-Way ANOVA [F(3, 10) = 0.4581] followed by Fisher’s LSD, *p < 0.05 to NT-MSC. (F) No significant changes in hippocampal Ube3aYfp transcript expression were found following either ZF-MSC IC or ZF-MSC CM treatment. One-Way ANOVA [F(3, 10) = 2.409] followed by Fisher’s LSD, *p < 0.05 to NT-MSC. (G) No significant changes were found in midbrain Ube3aYfp transcript expression following either ZF-MSC IC or ZF-MSC CM treatment. One-Way ANOVA [F(3, 10) = 2.035] followed by Fisher’s LSD, *p < 0.05 to NT-MSC. (H) No significant changes were found in cerebellar Ube3aYfp transcript expression following either ZF-MSC IC or ZF-MSC CM treatment. One-Way ANOVA [F(3, 10) = 1.560] followed by Fisher’s LSD, *p < 0.05 to NT-MSC. Error bars indicate ± standard error of the mean.
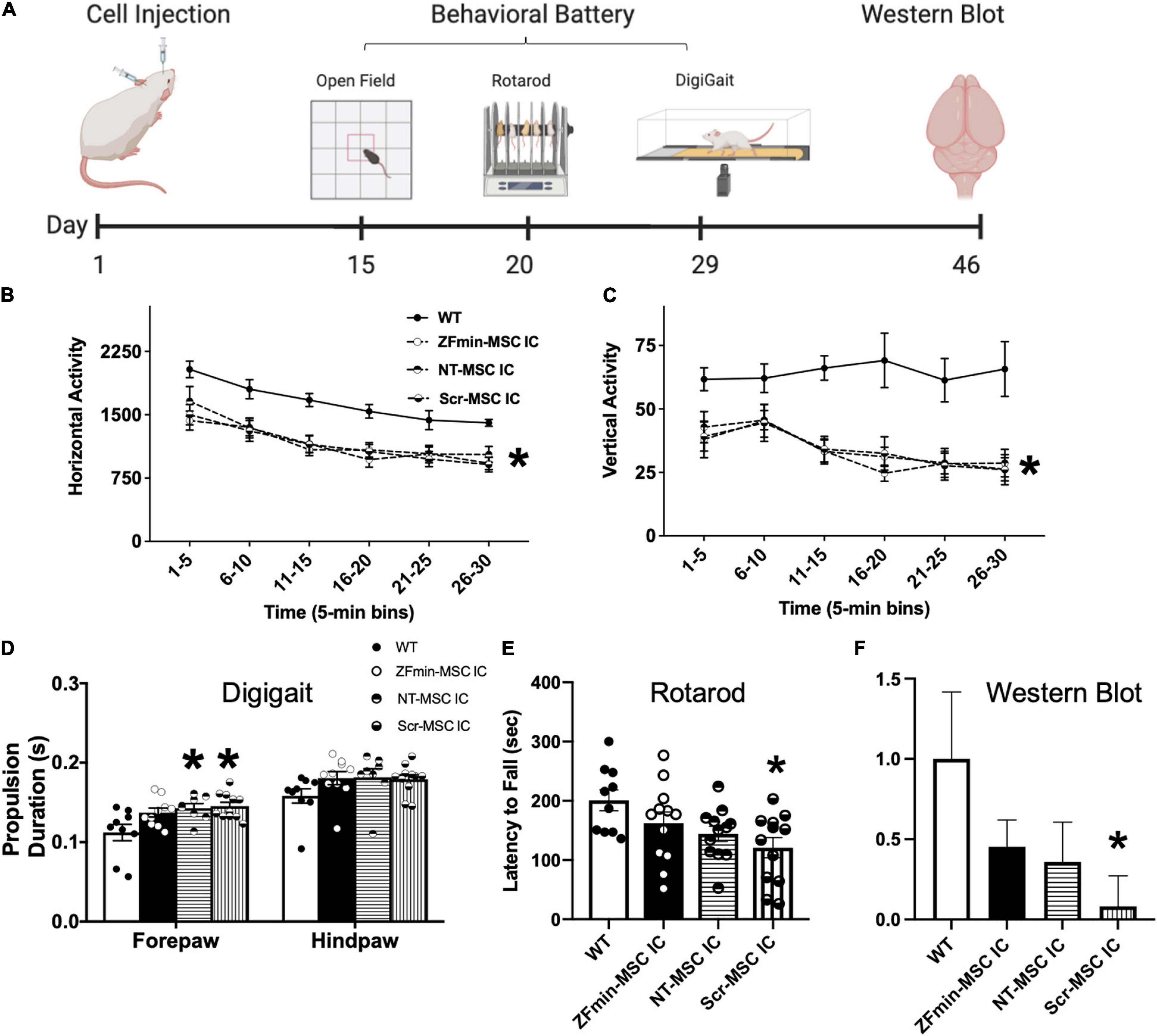
Figure 6. ZF-MSC improves subtle metrics of motor behavior but not gross motor ambulation. (A) Schematic of the timeline of cell injections, recovery time, tailored motor behavioral battery for large and small effects in the motor domain in Angelman Syndrome (AS) mice, and tissue collection. (B) Ube3amat–/pat+ mice exhibited typical, robust deficits in horizontal activity and (C) vertical activity compared to WT mice. No significant differences were found in locomotion in a novel open field assessing the horizontal and vertical activity using beam breaks in a novel arena. All AS groups, NT-MSC and Scr-MSC and ZF-MSC, regardless of treatment, were different from WT in horizontal activity and vertical activity. Analyses include both males and females. Mean ± standard error of the mean (SEM) is shown. *p < 0.05, two-way repeated-measures ANOVA (B) [F(4, 53) = 6.286], p < 0.001; (C) [F(4, 53) = 8.475]; p < 0.0001, the main effect of the treatment group followed by Sidak’s multiple comparison test. (D) Abnormal gait has been shown in Ube3amat–/pat+ mice and rats. While treadmill walking, the Scr-MSC and NT-MSC treated Ube3amat–/pat+ mice demonstrated significantly slower forepaw propulsion as compared to WT-littermates. Two-Way ANOVA [F(3, 34) = 3.949], p < 0.02; followed by Sidak’s Multiple Comparison Test. (E) Scr-MSC IC treated Ube3amat–/pat+ mice had significantly lower latencies to fall off the rotarod compared to WT, corroborating the finding in 6B. Two-Way ANOVA [F(3, 42) = 3.798], p < 0.02, a Sidak’s Multiple Comparison test could not confirm that performance of ZFmin-MSC IC animals was different from WT or control groups (p > 0.05). (F) Western Blot analysis of Ube3amat–/pat+ AS mice showed a significant difference in UBE3A protein expression in NT-MSC and Scr-MSC negative controls compared to WT mice, but not in ZFmin-MSC treated mice. One-Way ANOVA [F(3, 41) = 2.092] followed by Fisher’s LSD, *p < 0.05 from WT. Error bars indicate ± standard error of the mean. Dots indicate individual mice. n = 9–12 animals per group.
Zinc Finger-Mesenchymal Stem/Stromal Cell Intracranial Injections
Prior to surgery, rodent heads were shaved. Mice were anesthetized with isoflurane (2% in O2) and placed into a stereotaxic frame (Stoelting, Wood Dale, IL, United States) with ear bars for positioning. The head was aseptically cleaned with betadine and wiped clean with alcohol. A small incision was made in the scalp to allow visualization of the skull, with 1 mm burr holes at −1.5 mm AP and ± 1.25 mm ML relative to bregma. A total of 5 × 105 ZF-MSC or NT-MSC were injected anterior to the hippocampus at a depth of −1.75 using a Hamilton syringe at a rate of 0.5 μl/min with a total volume of 2 μl per side. After waiting an additional 5 min after injection, the burr hole was filled with bone wax and the incision was sutured with 6-0 silk.
Zinc Finger-Mesenchymal Stem/Stromal Cell Cisterna Magna Injections
Mice were prepared in a similar manner as noted above. Ear bars were raised until the skull of the mouse was angled at ∼35–40 degrees. A small incision was made at the base of the skull to allow visualization of the CM. The surrounding muscle anterior to the CM was carefully removed to visualize the base of the skull. The Hamilton syringe was placed at the same angle as the skull and was slowly lowered into the CM space with 1 × 106 cells infused at a rate of 0.5 μl/min with a total volume of 4 μl. After waiting an additional 1 min after injection, the incision was sutured closed with 6-0 silk.
In vivo Imaging Study
Mouse MSCs were transduced by a lentiviral vector carrying the luciferase gene (pCCLc-MNDU3-Luc-PGK-EGFP-WPRE), which allows cells to be visualized in the brains of living mice over time as previously described (Pollock et al., 2016). Bioluminescence was detected in anesthetized animals via IVIS Spectrum (Perkin Elmer, Waltham, MA, United States) (Perkin Elmer) 20 min post-intraperitoneal luciferin injection (3 mg/mouse, XenoLight D-Luciferin K+ Salt, Perkin Elmer, Catalog# 122799). Mice were imaged on post-op day 7 prior to the endpoint of the study.
Molecular Analysis
Tissue Extraction
On the last day of study, following cervical dislocation and rapid decapitation brains were rapidly extracted. The cerebral cortex, hippocampus, cerebellum, and midbrain were placed into 1.5 ml tubes and quickly frozen in liquid nitrogen. Tissue was homogenized using a tissue grinder (Kimble Chase, Vineland, NJ, United States, Kontes Pellet Pestle Motor, 749540-0000) and separated for genomic DNA, RNA, or protein.
Quantitative PCR Analysis
Tissue was immersed in 350 μl of TRIzol (Zymo Research, Catalog #R2050-1-200) and sonicated using a Dismembrator Sonicator (Thermo Fisher Scientific, Waltham, MA, United States) at setting 3 for 10 s. Sonicated tissue was then centrifuged at 10,000 × g relative centrifugal force (rcf) and supernatant was removed and incubated with an equivalent volume of 100% molecular grade ethanol, then RNA was extracted following Direct-Zol RNA Extraction (Zymo Research, San Diego, CA, United States, Catalog #R2053). cDNA was generated using RevertAid Random Hexamer (Thermo Fisher Scientific, Waltham, MA, United States, Catalog # K1691). Ten nanograms of cDNA was probed with TaqMan Universal PCR Master Mix (Thermo Fisher Scientific, Waltham, MA, United States, Catalog #4304437) including probes for Ube3a-Yfp (Thermo Fisher Scientific, Waltham, MA, United States, Catalog #10344-1-AP) and mGAPDH (Thermo Fisher Scientific, Waltham, MA, United States, Catalog MA1-16757) on a StepOne Plus (Applied Biosystems, Foster City, CA, United States). Ube3a-Ats qPCRs were assessed with probes for the 5′ Ube3a-Ats (Ube3a-Ats 5′ F – ACAGAACA ATAGGTCACCAGGTT and Ube3a-Ats 5′ R – AAGCAAGAC TGTTCACCTCAT) and 3′ Ube3a-Ats (Ube3a-ATS 3′ F – CCAA TGACTCATGATTGTCCTG and Ube3a-Ats 3′ R – GTGAT GGCCTTCAACAATCTC). Data are presented as 2–ΔΔCt normalized to Scr-MSC control group.
Western Blots
Protein was extracted via a standard protein isolation protocol using Pierce RIPA Buffer (Thermo Fisher Scientific, Waltham, MA, United States, Catalog #89901) with Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific, Waltham, MA, United States, Catalog #78442) and then quantitated using a Bicinchoninic acid assay protein kit (EMD Millipore, Burlington, MA, United States, Kit 71285-3). Forty micrograms of protein lysate was loaded into wells of 4–20% Stacking TGX Gels (BioRad, Hercules, CA, United States, Catalog #4561096EDU), were electrophoresed for 3 h at 100 V, then transferred onto prepared Invitrolon PVDF membrane (Thermo Fisher Scientific, Waltham, MA, United States, Catalog LC2005) and left overnight at 30 V. Membranes were blocked with 10% SEABLOCK salmon sperm (Thermo Fisher Scientific, Waltham, MA, United States, Catalog #37527). The membrane was then transferred to a solution containing rb-GFP (1:1,000, Novus Biologics, Centennial, CO, United States, Catalog #NB600 308) and rb-β-Tubulin (1:2,000, Thermo Fisher Scientific, Waltham, MA, United States, Catalog PA5-26039) with 5% SEA BLOCK in 1% TBST at room temperature for 2 h. Membranes were washed with TBST three times before incubation with 1:2,000 donkey rb-IgG (H + L) 800CW (LI-COR, Lincoln, NE, United States, Catalog #926-32213) and donkey ms-IgG (H + L) 680IR (LI-COR, Lincoln, NE, United States, Catalog #926-68070) in 5% SEABLOCK in 1% TBST for 1.5 h at room temperature. Gels were washed twice in 1% TBST and stored in TBS at 4°C. Bands were imaged using an Odyssey CLx (LI-COR, Lincoln, NE, United States). Optical density of western blots was quantified using ImageJ (NIH, Bethesda, MD, United States). Data are presented as relative percentages against a Ube3amatYfp/pat+ control.
Imaging
Tissue Sectioning
Mice used for immunohistochemistry (IHC) were euthanized using CO2 and transcardially perfused with 0.1 M PBS and fresh, ice-cold 4% paraformaldehyde [Avantor (J. T. Baker), Radnor, PA, United States, Catalog S898-07] at pH7.4. All brains were extracted and post-fixed in 4% paraformaldehyde for 36–48 h at 4°C and transferred to 30% sucrose in 0.1 M PBS for 48 h at 4°C. The brains were then cryopreserved in isopropanol on dry ice for 5 min and subsequently stored at ≤−80°C. Serially sagittal sections at 30 μM were obtained using a cryostat (Thermo Fisher Scientific, Waltham, MA, United States), starting from the midline and continuing laterally to a depth of ∼2,700 μm (15 sections in 6 wells), and were stored prior to labeling in a 0.02% sodium azide and 0.1 M PBS solution at 4°C.
Co-localization Imaging
Sagittal sections were incubated in a 10% SEABLOCK + 0.1% TBST blocking solution for 1 h prior to incubation with 1:500 rb-mCherry (abcam, Boston, MA, United States, Catalog #Ab167543) and 1:1,000 gp-NeuN (Sigma-Aldrich, St. Louis, MO, United States, Catalog #AB90) at 4°C overnight on gentle agitation. Tissue was then washed with TBST three times for 5 min before incubation with 1:1,000 goat rb-594 and 1:1,000 goat gp-647 AlexaFluor secondary antibodies (Thermo Fisher Scientific, Waltham, MA, United States, Catalog #A111012 and A21450), and then labeled with 1:1,000 Hoechst (Cell Signal Technology, Beverly, MA, United States, Catalog #4082) for nuclear visualization for 5 min. Tissue was then washed with TBST 2× prior to a final TBS 1× wash before mounting and coverslipped with Fluoromount (Sigma-Aldrich, St. Louis, MO, United States, Catalog #F4680-25ML).
3,3′-Diaminobenzidine Labeling
Sagittal sections were labeled with ImmPACT DAB Peroxidase Substrate (Vector Labs, Burlingame, CA, United States, Catalog #SK-4150), utilizing the Vectastain ABC Kit (Vector Labs, Burlingame, CA, United States, Catalog #HP1-26), following the manufactures protocol. Tissues underwent endogenous peroxidase quenching using a 0.3% hydrogen peroxide solution in water for 30 min, followed by immersion in a 10% blocking solution in 0.1% PBST + SEABLOCK Blocking Buffer (Thermo Fisher Scientific, Waltham, MA, United States, Catalog #37527), for 1 h, followed by incubation in primary antibody 1:1,000 rb-GFP (Novus Biologicals, Centennial, CO, United States, Catalog #300-608) and incubated overnight at 4°C. On the second day, tissues were immersed in a biotinylated Goat ms-IgG secondary antibody (Vector Labs, Burlingame, CA, United States, Catalog #BA-9200) solution at a concentration of 1:200 with incubation for 1 h, followed by Vectastain ABC reagent (Vector Labs, Catalog # HP1-26) incubation for 30 min, and concluded with immersion into ImmPACT DAB Peroxidase Substrate (Catalog #SK-4150) at the manufacturers recommended concentration for 8 min. In between each step all tissues were washed using PBST (0.1% Triton) for ∼15 min. Serial sections were mounted onto uncharged slides and coverslipped using Permount Mounting Medium (Thermo Fisher Scientific, Waltham, MA, United States, Catalog #SP15-100).
Image Acquisition and Analysis
Images of fluorescent and DAB labels were captured using a Zeiss Observer Z1. Structural specific imaging was performed at 20× for the prefrontal cortex, cerebral cortex, striatum, hippocampus, and cerebellum. Co-localization imaging was completed using the apotome with a 40× objective. Whole-brain imaging was performed at 5× and stitched together using ZEN 2.6. All images were analyzed using ImageJ (NIH, Bethesda, MD, United States). MSC and ZF volume of distribution was calculated using Cavalieri’s volume estimation. The average intensity of YFP was measured in the hippocampus. Images for co-localization were taken with a 40×/0.8 aperture with an inserted ApoSlider at CA1 or CA3. Yfp/NeuN channels were exported and analyzed using a Mander’s Co-Localization Analysis followed by a Costes’ Thresholding through the JaCoP Image J plugin (Bolte and Cordelières, 2006).
Mouse Behavior
Mice (n = 46) were housed in Techniplast cages (Techniplast, West Chester, PA, United States). Cages were housed in ventilated racks in a temperature (68–72°F) and humidity (∼25%) controlled colony room on a 12:12 light/dark cycle. Standard rodent chow and tap water were available ad libitum. In addition to standard bedding, a Nestlet square, shredded brown paper, and a cardboard tube (Jonesville Corporation, Jonesville, MI, United States) were provided in each cage for enrichment. The order and age of testing were as follows: (1) Open field at 8 weeks of age, (2) Rotarod at 9 weeks of age, and (3) DigiGait at 10 weeks of age.
Open Field
General exploratory locomotion in a novel open field arena was evaluated as previously described (Ku et al., 2016; Berg et al., 2018). Briefly, each subject was tested in a VersaMax Animal Activity Monitoring System (Accuscan, Columbus, OH, United States) for 30-min in a ∼30 lux testing room.
Rotarod
Motor coordination, balance, and motor learning were tested with an accelerating rotarod (Ugo Basile, Gemonio, Italy) as previously described (Williams, 2010). Mice were placed on a rotating cylinder that slowly accelerated from 5 to 40 revolutions per min over 5 min. Mice were given three trials per day with a 60-min inter-trial rest interval and tested for 3 consecutive days for a total of nine trials. Performance was scored as latency to fall off the cylinder with a maximum latency of 5 min.
Gait Analysis
Treadmill gait analysis was performed using the DigiGait™ system (Mouse Specifics Inc., United States). Mouse paws were painted with non-toxic red food coloring to augment dark green paw tattoos that generated conflict in DigiGait™ analysis one minute prior to introduction to the walking chamber to reliably capture the entire paw surface area. For each mouse, videos of ∼5 sec duration of all sessions were analyzed using the DigiGait™ Imaging and Analysis software v12.2 (Mouse Specifics Inc., United States). Contrast filters were determined on a mouse-by-mouse case to facilitate consistent recognition of all four paws. All analysis was conducted in a single session by experimenter blind to genotype. Data was averaged between left and right paws for fore and hind paws.
Rhesus Monkeys
All animal procedures conformed to the requirements of the Animal Welfare Act and protocols were approved prior to implementation by the Institutional Animal Care and Use Committee at the University of California, Davis. Three young rhesus monkeys (Macaca mulatta) (∼3 months of age; 1 kg) were included in these studies. Autologous rhesus monkey bone marrow-derived MSCs (rhMSC) were obtained and cells were grown in culture and transduced according to established published protocols (Figure 7A; Lee et al., 2004, 2006). Luciferase expression was confirmed with bioluminescence imaging (BLI) prior to in vivo administration (IVIS ® Spectrum in vivo Imaging System with Living Image Software; PerkinElmer). A total of 1 × 106 rhMSC expressing firefly luciferase were administered to each of the telazol-sedated animals (5–8 mg/kg) using an established IT approach (0.5 ml CSF removed using a 25 g needle and 3 ml syringe, then the syringe was changed to inject ∼0.5 ml rhMSC slowly) under aseptic conditions (Hordeaux et al., 2019). BLI was immediately performed post-injection of rhMSC and after the intravenous administration of D-Luciferin (100 mg/ml) according to established methods (Figure 7B; Tarantal et al., 2009; Tarantal and Lee, 2010). Animals were monitored throughout the day then daily physical signs were assessed with peripheral blood sample collection (∼2–3 ml) and body weight weekly [complete blood counts, chemistry panels; all within normal limits (data not shown)]. At 3 weeks post-administration animals were sedated with ketamine, blood samples and CSF collected, then euthanized with an overdose of pentobarbital for tissue harvests according to established methods. Multiple sections of the brain (right and left frontal, parietal, temporal, occipital lobes; hippocampus; right and left cerebellum; midbrain) and spinal cord (cervical, thoracic, and lumbar) were collected and placed in sterile tubes and quick frozen over liquid nitrogen and stored at ≤−80°C until assay. The presence of ZFs was evaluated following the same western blot techniques described above.
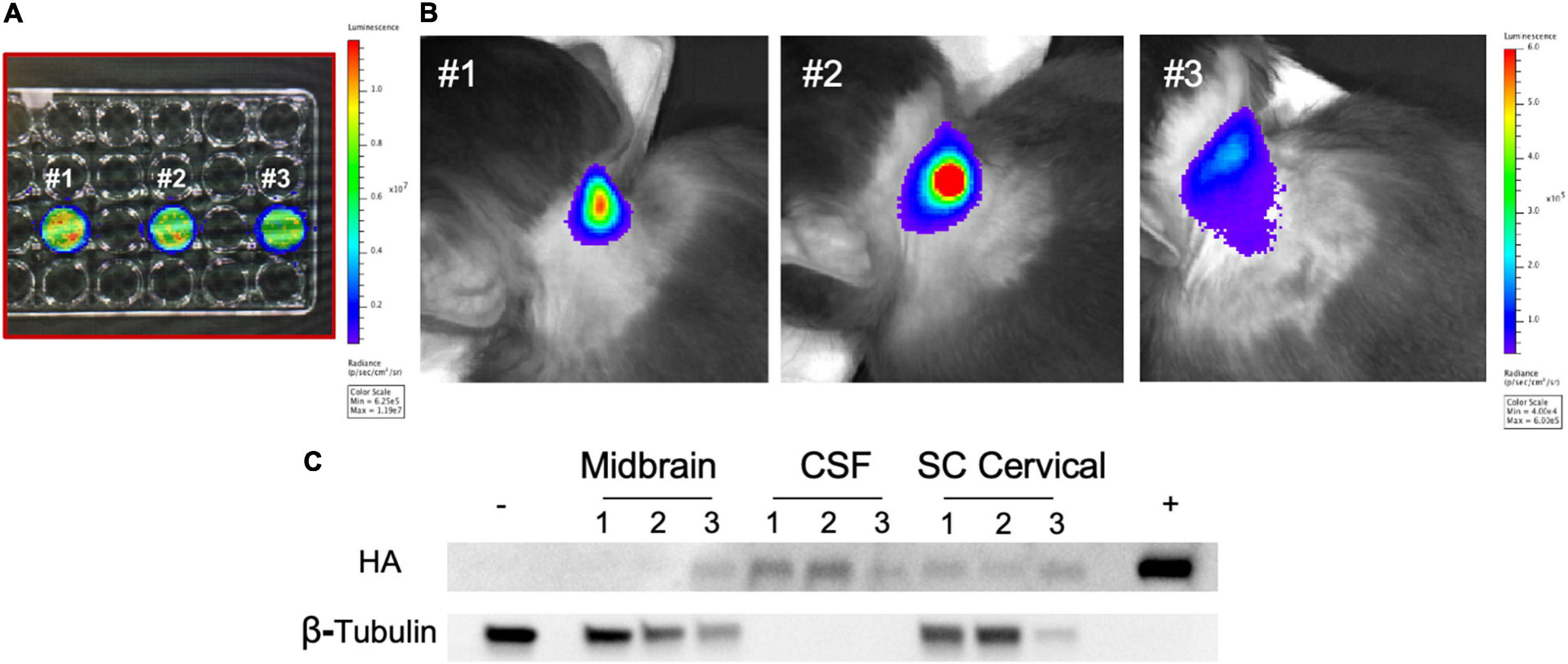
Figure 7. Intrathecal administration of autologous rhMSC in young rhesus monkeys. (A) Autologous rhMSC were collected per standard protocol and cells transduced with a lentiviral vector expressing firefly luciferase. rhMSC were confirmed to express luciferase by bioluminescence imaging (BLI) prior to intrathecal injection. Numbers reflect transduced rhMSC from monkey #1, monkey #2, and monkey #3. (B) BLI was performed immediately following intrathecal administration of rhMSC expressing firefly luciferase according to established protocols. The location of the firefly luciferase-expressing autologous rhMSC post-injection are shown in representative images from each rhesus monkey, consistent with findings in other cell and gene transfer studies where BLI has been used (Tarantal et al., 2009; Tarantal and Lee, 2010). (C) Western blot shows the presence of zinc finger (ZF) 3 weeks following injection. ZF protein was delectable in the CSF (three animals), midbrain (one animal), and spinal cord (two animals). Historical whole brain lysate from non-treated rhesus monkeys was used as a negative control. ZF-MSC lysate used as a positive control. Numbers reflect animal number as described in (A).
Statistics
All statistical analysis was performed using GraphPad Prism 7 with an alpha level equal to 0.05. All molecular and histological data was analyzed using an analysis of variance or Student’s t-test where appropriate to assess differences between groups. When appropriate, a Fisher’s Least Significant Difference post hoc test was also performed.
Results
Engineered Mouse Mesenchymal Stem/Stromal Cell Can Function as Biofactories for Zinc Finger and Transcription Activator-Like Effectors
The initial ZF protein construct contained an N-terminal maltose-binding protein for protein purification, the 10-aa transduction domain of the HIV-transactivator protein (TAT, residues 48–57) as a cell-penetrating peptide, a mCherry red fluorescent protein to aid in and visualization, a Hemagglutinin (HA) epitope tag for detection, and a SV40 nuclear localization signal to ensure nuclear delivery following cellular transduction. The TAT cell-penetrating peptide had been previously shown to deliver other proteins to the mouse brain following intraperitoneal injection (Gong et al., 2006; Wu et al., 2015). The C-terminus of the protein consisted of the ZF array and KRAB repressor domain. We replaced the maltose binding protein with an IgK Signal Peptide as a secretion peptide and altered the original TAT peptide with a furin-resistant TAT (TATk) for improved protein stability (Flinterman et al., 2009) and subcloned into a lentivirus compatible vector This specific orientation of IgK-TATk-mCherry-HA-NLS-ZF-KRAB would allow for active secretion of the fully translated protein into the extracellular space of a cell, followed by uptake into neighboring cells through the TAT peptide, and then localization to the nucleus to silence the Ube3a-Ats (Figure 1A). We transduced mouse bone marrow derived MSCs with this ZF secretion vector (ZF-MSC) and found secretion of the full length 67-kDa ZF protein in the ZF-MSC-conditioned media (Figure 1B). We incubated ZF-MSC-conditioned media with Neuro2A cells and observed co-localization of mCherry-tagged ZF in recipient cells within 24 h (Figure 1C, right panel). Additionally, engineered MSCs were able to secrete ATF of different sizes such as a smaller 62-kDa ZF and a 120 kDa TALE (Supplementary Figures 1A,B). We evaluated alterations in MSC phenotype following transduction and found that both a ZF-MSC and a scramble ZF MSC (Scr-MSC) demonstrated typical MSC markers as assessed by flow cytometry and did not affect proliferation when compared to non-transduced MSC (NT-MSC) controls (Figures 1D,E and Supplementary Figures 2, 3).
In vitro Functional Validation of Secreted Zinc Finger Protein
Our designed ZF binds to the putative Ube3a-Ats promoter and represses expression of this long non-coding antisense RNA, allowing for expression of the intact paternal Ube3a gene (Figure 2A). We evaluated the biological functionality of an MSC-mediated secreted ZF in a series of in vitro assays. The therapeutic potential of MSCs is thought to be mediated by two mechanisms of action—microtubule conjugation with neighboring cells to allow for direct transfer of cargo or secretion of bioactive molecules that act in a paracrine manner (Liang et al., 2014). The paracrine mechanism of action was evaluated through co-incubation of engineered MSCs with recipient Neuro2A mouse neuroblastoma cells in Transwell Assays (Figure 2B). Both our ZF-MSC and an epitope-free, ZFmin-MSC variant, demonstrated reduction in Snrpn expression, the proximal gene to where the ZF binds, following co-incubation with Neuro2A cells. We observed that the Neuro2As treated with the purified protein version of ZF demonstrated a significant 28.6% reduction of Snrpn transcript expression 14 h following treatment compared to a NT-MSC conditioned media via RT-qPCR (Figure 2C). This effect was ameliorated at 24 h (Figure 2D) and 48 h (Figure 2B) compared to NT-MSC control. In comparison, we did not observe a significant change in Snrpn expression following a 24 h incubation with either ZF variant (Figure 2D). At 48 h, a significant 15% reduction in Snrpn was observed in ZFmin-MSC treated cells but no significant changes were detected with ZF-MSC (Figure 2E). At 72 h, significant Snrpn reduction was observed in ZF-MSC (24.7%) and ZFmin-MSC (20.2%) treated cells relative to the NT-MSC control (Figure 2F).
Next, we evaluated secreted ZF functionality in primary cortical cultures from the Ube3a+/yfp reporter mouse model. The paternal Ube3a is fused to a yellow fluorescent protein (Yfp) reporter and is silenced in mature neurons (Dindot et al., 2008). We incubated conditioned media from ZF-MSC, ZFmin-MSC, or purified ZF in primary cortical neurons harvested from E15.5 Ube3a+/yfp pups for 24 h (Figure 2E). Immunocytochemistry (ICC) of treated primary cortical neurons revealed an increase of YFP expression in all treatment groups compared to Scr-MSC conditioned media and no significant differences in YFP expression between purified ZF treatment and ZF-MSC variants following a 24 h incubation with conditioned media (Figures 2F,G). Western blot analysis demonstrated increased UBE3A-YFP in treated samples (Figure 2H). Together, these data demonstrate the functionality of secreted ZF from MSCs in both immortalized and primary mouse cell lines to activate paternal Ube3a expression.
Robust Protein Uptake and Biological Effect of Secreted Zinc Finger Protein
We evaluated our platform in vivo to understand the kinetics and distribution of a cell secreted ZF across different routes of administration. Initially, a total of 1 × 106 ZF-MSC were injected intracranially (IC) into the striatum or via the CM in wild-type (WT) FVB mice. We observed injected MSCs at the site of IC injection (Figure 3A) and presence of secreted ZF protein in the extracellular space of IC and CM injections (Figures 3A–C). ZF-MSC IC protein appeared to distribute from the injection site as measured from both the dorsal (cortex) and ventral (striatum) plane. A Mander’s Colocalization Coefficient Analysis followed by a Costes’ automatic thresholding revealed a gradient-specific decline of protein uptake in NeuN+ cells that are more distally anterior and posterior from the injection site (Figures 3D,E). Interestingly, ZF-MSC CM protein also demonstrated a gradient-dependent distribution of ZF protein uptake from the site of CM injection (Figure 3F).
Following the confirmation of in vivo secretion of ZF protein, 1 × 106 ZF-MSC were bilaterally injected intracranially dorsal to the region of interest in AS, the hippocampus, or via the CM in the Ube3a+/yfp reporter mouse. BLI demonstrated that transduced MSCs persisted at least 1 week following injection (Figure 4A). For IC delivery, IHC analysis revealed that ZF-MSCs were observed along the needle tract and the white matter in the corpus callosum in IC-injected mice (Figure 4B and Supplementary Figure 4). MSCs were found in the third ventricle and the cerebellum (Supplementary Figure 4). In contrast, MSCs delivered via the CM were observed along the surface of the cortex, cerebellum, spinal cord, and hypothalamus of the mouse brain (Supplementary Figure 5). Secreted ZF protein was observed in NeuN+ cells within the CA1 and CA3 regions of the hippocampus following both routes of administration (Figure 4C). A Mander’s Colocalization Coefficient Analysis followed by a Costes’ automatic thresholding revealed that IC delivery resulted in 46.8% of CA1 and 39.1% of CA3 NeuN+ areas co-labeled with ZF protein as compared to 7.6% and 9.8%, respectively, in mice administered MSC via CM delivery (Figures 4D,E).
Zinc Finger-Mesenchymal Stem/Stromal Cell Activates Paternal Ube3aYfp Expression in the Ube3a+/Yfp Mouse
Yellow fluorescent protein activation was observed in morphologically distinct pyramidal neurons in the hippocampus of ZF-MSC administered IC mice using 3,3′-Diaminobenzidine (DAB) IHC 1 week post-delivery (Figure 4F). Relative YFP mean intensities of IC injected ZF-MSC treated mice demonstrated a significant 27.8% increase (Figure 4G) in CA1 and a significant 16.1% increase (Figure 4H) in CA3 of the paternally imprinted UBE3A-YFP protein in ZF-MSC treated Ube3a+/yfp mice when compared to the NT-MSC controls. No effect was observed in the CM group at the 1-week time point (Figures 4G,H). We observed a positive correlation between ZF co-localization and YFP intensity in CA1 (Figure 4I), and CA3 (Figure 4J).
Paternal Ube3aYfp activation was further evaluated in discrete brain regions through molecular assays. We observed a route of administration-specific divergence in which neuronal subregions showed alterations in Ube3aYfp expression. Ube3a+/yfp mice administered ZF-MSC IC or ZF-MSC CM did not demonstrate a difference in UBE3A-YFP protein expression in the cortex compared to Scr-MSC IC groups (Figure 5A). In the hippocampus, there was a significant 9.9% and 8.5% increase in UBE3A-YFP protein of ZF-MSC IC and ZF-MSC CM, respectively (Figure 5B). In the midbrain, there was a significant 27.6% and 19% increase in the UBE3A-YFP protein of ZF-MSC IC and ZF-MSC CM treated mice, respectively (Figure 5C). In the cerebellum, there was a significant 25.5% increase in UBE3A-YFP protein of ZF-MSC CM treated mice, but no observable effect in ZF-MSC IC mice (Figure 5D). No significant effects were observed in Yfp transcript expression (Figures 5E–H). This transcriptional data was further corroborated by no observable changes in either the 3′ or 5′ Ube3a-Ats transcript (Supplementary Figure 6). Together these data demonstrate in vivo activation of Ube3aYfp protein, but not transcript 3 weeks after injection.
Zinc Finger-Mesenchymal Stem/Stromal Cell Attenuates Motor Deficits in the Ube3amat–/pat+ Angelman Syndrome Mouse
Following activation of paternal Ube3aYfp in the Ube3a+/yfp mice, we utilized IC delivery of ZFmin-MSC for behavioral assessments in the Ube3amat–/pat+ AS mouse model (Jiang et al., 1998). Motor deficits have been consistently reported across mouse models of AS, such as dysfunction on the rotarod and reduced activity in a novel arena (Huang et al., 2013; Born et al., 2017; Sonzogni et al., 2018), thus we focused on a motor phenotyping battery for functional rescue. Six-week-old Ube3amat–/pat+ mice underwent bilateral IC injection of 1 × 106 ZFmin-MSC and were allowed to recover for 2 weeks following surgery before undergoing a motor-tailored behavioral battery (Figure 6A). All AS groups regardless of treatment differed from WT in horizontal activity (Figure 6B) and vertical activity (Figure 6C), a hallmark of AS pathology (Adhikari et al., 2021). In gait analysis, NT-MSC and scrambled ZF MSC) treated Ube3amat–/pat+ mice showed deficits in forelimb propulsion as time in the propulsion phase was significantly increased compared to WT littermates (Figure 6D) whereas ZFmin-MSC treated mice were indistinguishable from WT mice. Scr-MSC treated mice also exhibited behavior that is typical of Ube3amat–/pat+ mice and rats with reduced latencies to fall on an accelerating rotarod assay compared to WT-littermates (Huang et al., 2013; Born et al., 2017; Berg et al., 2020). Both NT-MSC and ZFmin-MSC treated mice demonstrated an intermediate effect; while there was an overall genotypic effect (Figure 6E), performance was not different from WT or Scr-MSC groups. We observed a significant reduction in UBE3A protein in Scr-MSC treated mice as compared to WT littermates; however, ZFmin-MSC and NT-MSC were not significantly different from either of those groups (Figure 6F).
Young Rhesus Monkeys Demonstrate Presence of Secreted Zinc Finger
Autologous rhMSC were co-transduced to express firefly luciferase and secrete ZF (ZF-rhMSC) (Figures 7A,B). ZF-rhMSC (1 × 106) were injected under aseptic conditions using an established IT approach and were well-tolerated for the duration of the 3-week study. CBCs and chemistry panels within normal limits (data not shown). Tissues were collected 21 days post-administration and assessed for the presence of ZF protein. Secreted ZF protein was detected in the CSF (2 of 3 animals), midbrain (1 of 3 animals), and spinal cord (3 of 3 animals) (Figure 7C).
Discussion
Mesenchymal stem/stromal cells have been used as a delivery system for bioactive molecules in cross correction approaches (Meyerrose et al., 2010; Damasceno et al., 2020) and gene modifying molecules such as shRNAs (Olson et al., 2012). Here, we adapted the innate paracrine abilities of MSCs to secrete artificial transcription factors of various sizes that remain bioactive in vivo. We report the use of MSCs to secrete a bioactive ZF to alter Ube3a expression in AS mouse models and demonstrate detectable distribution in midbrain and spinal cord of rhesus monkeys. We sought to evaluate if the secretable ZF protein would retain its ability to distribute widely in the brain using multiple routes of administration (surgical IC, CM, or IT).
We report co-localization of secreted ZF protein with mature neurons in the mouse hippocampus 1 week following either an IC injection that was proximal to the hippocampus or a more distal CM injection. Direct ZF-MSC injection into the parenchyma of the mouse brain demonstrated a passive diffusion of secreted protein from the bolus of cells that was likely taken up by neighboring cells. Interestingly, secreted ZF was detectable in the deeper, midbrain in the mouse and monkey. ZF-MSCs were detectable within the spinal cord of ZF-MSC treated mice upon CM delivery and thus likely a source of ZF protein in the CSF. This was also shown by detection of the ZF protein within the spinal cord and CSF of rhesus monkeys. It is not well understood how a TAT-fused peptide is able to penetrate deeper, subcortical regions that are distal from the arachnoid space as the flow of CSF is canonically thought to originate in the choroid plexus and then flow into the lateral ventricles, the third ventricle, the cisterns, the arachnoid space, and then empty into the arachnoid villi (Brinker et al., 2014). Interestingly, Papisov et al. (2012) demonstrated that lumbar IT injection of large macromolecules rapidly distribute into the CSF and are taken up into the tissue, potentially aided by pulsatile movements of arteries in the leptomeningeal space. Mazur et al. (2019) demonstrated that IT administration of macromolecules such as antisense oligonucleotides penetrate subcortical regions such as the hippocampus and thalamus of the brain more slowly compared to cortical spaces and hypothesized that this may be mediated by exchange of proteins into the perivascular spaces that have deeper brain access (Barua et al., 2014). We and others have previously shown that the inclusion of a TAT-peptide allows for robust penetration of recombinant proteins along the brain parenchyma (Schwarze et al., 1999; Kilic et al., 2002, 2003; Doeppner et al., 2010; Ye et al., 2011; Wu et al., 2015) and this likely enhances the ability for a secreted protein to have a broad distribution along the neuroaxis. Furthermore, MSCs are well known to have a vast secretome that is mediated through the release of exosomes. The present cell delivery system is designed to secrete naked protein, however, further evaluation of the potential contribution of exosomal delivery of ATFs is of interest for future investigation.
Zinc finger-mesenchymal stem/stromal cell secreted protein and purified ZF protein (Bailus et al., 2016) demonstrated differing pharmacokinetic profiles in vitro. Purified ZF induces an immediate reduction of Snrpn expression that quickly tapers off due to the relatively short half-life of ZF proteins (Ramakrishna et al., 2013; Bailus et al., 2016). In comparison, ZF-MSC are able to induce a sustained reduction of Snrpn within 48 h of incubation. This delayed effect is likely due to the time necessary for sufficient ZF protein to be produced and secreted by MSC. We observed a significant increase in hippocampal UBE3A-YFP in Ube3a+/yfp mice administered ZF-MSC by the IC route within 1 week of injection. However, no effect was observable in the CM group. We found that the amount of ZF co-localization with hippocampal neurons was strongly correlated with increases in UBE3A-YFP, suggesting that sufficient exposure to ZF protein is necessary to elicit a biological effect—in agreement with our in vitro and in vivo data that demonstrates a slower ZF accumulation in regions that are more distal to the CM.
We also observed a divergence in the brain regions that are amenable to alterations of gene expression concerning the different administration routes studied. At 3 weeks, mice treated by either delivery route expressed more UBE3A-YFP in the midbrain and hippocampus, whereas only CM delivery of ZF-MSC resulted in the expression of more UBE3A-YFP in the cerebellum. It is interesting to note that ZF-MSCs are highly motile in the brain following either route of administration—a similar phenomenon that has been observed in other tissues (Chen et al., 2015; Argibay et al., 2017). Intracranial-delivered ZF-MSC appeared to travel along the corpus callosum toward CSF-rich spaces such as the lateral ventricles where the cells could be detected throughout areas in the brain such as the third ventricle and the cerebellum. CM delivered ZF-MSC were detectable in the spinal cord, cerebellum, and along the apical or basal regions of the cortex. The ability for ZF-MSCs to migrate and embed in other regions of the mouse brain may promote a localized accumulation of ZF in CSF-rich recesses such as the lateral ventricles to induce paternal Ube3aYfp activation in subcortical regions of the mouse brain. The ability for MSCs to travel beyond their site of injection may further potentiate their ability to act as roaming biofactories within the CNS. Data supports that a less invasive, non-surgical CM injection effectively delivers engineered MSC in local and distal regions of the brain. Additionally, this observation is confirmed in studies that tested the feasibility, safety, and tolerability of IT administration of ZF-rhMSCs in young rhesus monkeys. These cells were well-tolerated and did not result in any adverse events for the duration of the study. Importantly, secreted ZF protein was detectable within the CSF, midbrain, and spinal cord in rhesus. Previous examples in the gene therapy field indicate that transitioning directly from mouse studies to human clinical trials is fraught with species and technical limitations, including anatomical and physiological differences between brain structure and size. This concern necessitates the use of a larger primate species such as the rhesus monkey that simulates human anatomy and physiology. Our data support that our system allows for delivery of ZF to the brain in a safe, non-surgical manner that may allow for administration of engineered MSC into the CSF to promote a biological effect.
Movement disorders affect nearly every individual with AS (Tan et al., 2011; Wheeler et al., 2017; Keute et al., 2020). Previous work by Silva-Santos et al. (2015) demonstrated that 70% restoration of Ube3a at postnatal day 21 in an inducible Ube3a mouse resulted in only a partial recovery of rotarod performance, suggesting that there was a critical window of treatment at 21–42 days following birth. This was further corroborated by recent work by Wolter et al. (2020) showing prenatal intervention in Ube3a-Ats expression resulted in attenuation of disease phenotypes and restoration of Ube3a expression. In our study, ZF-MSCs were able to induce improvements in forepaw propulsion and latency to fall on the accelerating rotarod in Ube3amat–/pat+ mice when treated on post-natal day 42. Our results suggest that ZF-MSC treatment improved subtle motor control as mice were rapidly placing their paws onto the ground or improving muscular weakness as less time was required for forelimb strides, which was significantly elevated in Ube3amat–/pat+ relative to WT littermate controls. It is interesting to note that an intermediate effect was also observed for NT-MSC treated mice in the latency to fall on the accelerating rotarod task. MSCs secrete several favorable trophic factors including brain-derived neurotrophic factor and noggin that can improve phenotypic outcomes in mice (Lu et al., 2003). While our scrambled controls did not induce any improvements in accelerod, we cannot discount the effect that unaltered MSCs alone may have on motor phenotypes. Importantly, we only observed an intermediate effect in Ube3a expression in ZFmin-MSC following IC surgeries, suggesting that an epitope-free form of the ZF has a favorable pharmacodynamic profile for up to 29 days.
Previous work by our group and others suggest that the biologic effect of a KRAB-fused ATF is transient transcriptional repression (Gilbert et al., 2014) and is contingent on presence of the gene modifying protein. UBE3A has a half-life of ∼61 h (Olabarria et al., 2019). We did not observe a detectable presence of ZF-MSC 3 weeks following injection through either route of administration in mice, suggesting that injected MSCs were rapidly cleared from the brain parenchyma. Thus, the lack of Ube3aYfp transcriptional expression may be indicative of a tapering of the therapeutic effect as the ZF protein clears while elevated UBE3A-YFP protein expression persists for some time post-injection. Our current platform may benefit from continued advances in defining gene modifiers that induce durable epigenetic markers that result in sustained alterations in gene expression (Amabile et al., 2016; O’Geen et al., 2019; Nuñez et al., 2021). Temporal secretion of such gene modifiers that are capable of widespread coverage within the CNS through cellular biofactories may offer an ideal delivery vehicle for “hit-and-run” epigenetics.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
Animal studies were approved by the UC Davis Institutional Animal Care and Use Committee.
Author Contributions
PD, AT, JS, DS, and KF: conceptualization. PD, FF, AT, JS, DS, and KF: methodology. PD, JH, UB, RL, DC, JW, KT, MP, AA, NC, SP, HO’G, SL, AT, MM, CL, and KF: investigation. AT, JN, JS, and KF: resources. PD: writing—original draft. PD, JH, AT, JN, JS, DS, and KF: writing—review and editing. PD and JH: visualization. KF: supervision and project administration. AT, JN, DS, and KF: funding acquisition. All authors contributed to the article and approved the submitted version.
Funding
The studies described was supported by a CIRM Discovery Grant DISC2-09032, Diversity Supplement 3R24OD021606-02ZF, NIGMS Pharmacology T32 GM099608, NIH NCATS UL1 TR001860 and linked award TL1 TR001861 (mice); and a California National Primate Research Center pilot project award (rhesus monkeys; NIH P51-OD011107). The Dickenson’s Catalyst Fund, A Stewart’s and Dake Family Gift, HELP4HD International, WeHaveAFace.org, and philanthropic donors from the Huntington’s Disease community, including the Roberson family and Team KJ, also contributed to these studies. BLI was performed in rhesus monkeys with instrumentation obtained through a NIH S10 award to AT (S10-OD018102). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The Center for Molecular and Genomic Imaging at UC Davis for mouse figure generation and Biorender for general figure generation.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fnmol.2021.789913/full#supplementary-material
Supplementary Figure 1 | Transduced MSCs are able to secrete full-length zinc finger and transcription activator-like effector proteins, related to Figure 1. (A) Full-length 62-kDa ZF is detectable in conditioned media from ZF-MSCs but absent in isogenic NT-MSCs. Plasmid map indicates size of secretion ZF transgene and predicted protein size. (B) Full-length 120-kDa TALE is detectable in conditioned media from TALE-MSCs but absent in isogenic NT-MSCs. Plasmid map indicates size of secretion TALE transgene and predicted protein size.
Supplementary Figure 2 | Transduced mouse BM-MSCs demonstrate positive canonical MSC markers CD44b and negative for CD11b, CD34b, and CD106, related to Figure 1. (A) Isotype FITC negative control. (B) NT MSC, Scr-MSC, and ZF-MSCs are negative for immune marker CD11b. (C) NT MSC, Scr-MSC, and ZF-MSCs are negative for hemopoietic stem cell marker CD34b. (D) NT MSC, Scr-MSC, and ZF-MSCs are positive for MSC surface marker CD44b. (E) NT MSC, Scr-MSC, and ZF-MSCs are negative for vascular adhesion marker CD106.
Supplementary Figure 3 | Transduced mouse BM-MSCs demonstrate positive canonical MSC markers CD29b and Sca1 and negative for CD31, CD45, and CD105, related to Figure 1. (A) Isotype PE negative control. (B) NT MSC, Scr-MSC, and ZF-MSCs are positive for MSC surface marker CD29b. (C) NT MSC, Scr-MSC, and ZF-MSCs are negative for platelet endothelial adhesion molecular marker CD31. (D) NT MSC, Scr-MSC, and ZF-MSCs are negative for differentiated hemopoietic cell marker CD45b. (E) NT MSC, Scr-MSC, and ZF-MSCs are negative for vascular endothelial marker CD105. (F) NT MSC, Scr-MSC, and ZF-MSCs are enriched for mouse MSC marker Sca-1.
Supplementary Figure 4 | Intracranial injection of ZF-MSC demonstrated motility from the parenchyma, related to Figure 4. (A) NT-MSC IC was observable within the mouse brain one week following injection and detected moving towards the lateral ventricles along the corpus callosum (A1). The white box denotes inlet of an image in panel (A1). Dashed white lines indicate the presence of injected MSC due to distinct morphology in contrast to mature NeuN+ (red) neurons and nuclear Hoechst (blue) labeling. (B) ZF-MSC IC (green) was observable within the mouse brain following injection and detected moving along the corpus callosum towards the lateral ventricles (B1). A white box denotes an inlet of image in panel (B1). (C) ZF-MSCs IC were detectable across multiple regions of the brain, including the forebrain (C1), site of injection along the corpus callosum (C2), anterior to the hippocampus (C3), along the third ventricle–ventral (C4) and posterior (C5) to the hippocampus, in the visual cortex (C6), along with the cerebellum (C7,C8), and within the lateral ventricles (C9). Scale bar = 500 μm (A–C), 50 μm (A1,B1,C1–C9).
Supplementary Figure 5 | Cisterna magna injection of ZF-MSC demonstrated motility to the CSF one week following injection, related to Figure 4. (A) Cisterna magna (CM) injected ZF+ (green) were detectable along the somatosensory cortex (A1) and were distinct from surrounding NeuN+ (red) neurons and other Hoechst (blue) labeled cells. (B) ZF-MSC injected via the CM were detectable across multiple regions including the cortex (B1), cerebellum (B2–B4) and along hypothalamic regions (B5–B6). (C) ZF-MSC injected via the CM were detectable within the mouse spinal cord. Scale bar = 500 μm (A–C), 50 μm (A1,B–B6).
Supplementary Figure 6 | YFP transcript expression is not altered with response to ZF, related to Figure 5. (A) No observable changes in the 3′ Ube3a-Ats transcript was observed in the cortex, hippocampus, midbrain, or cerebellum in 3-weeks post ZF-MSC treatment in Ube3a+/yfp mice. (B) No observable changes in the 5′ Ube3a-Ats transcript was observed in the cortex, hippocampus, midbrain, or cerebellum in 3-weeks post ZF-MSC treatment in Ube3a+/yfp mice. Error bars indicate ± standard error of the mean. Dots indicate individual mice.
References
Adhikari, A., Copping, N. A., Beegle, J., Cameron, D. L., Deng, P., O’Geen, H., et al. (2021). Functional rescue in an Angelman syndrome model following treatment with lentivector transduced hematopoietic stem cells. Hum. Mol. Genet. 30, 1067–1083. doi: 10.1093/hmg/ddab104
Adhikari, A., Copping, N. A., Onaga, B., Pride, M. C., Coulson, R. L., Yang, M., et al. (2019). Cognitive deficits in the Snord116 deletion mouse model for Prader-Willi syndrome. Neurobiol. Learn. Mem. 165:106874. doi: 10.1016/j.nlm.2018.05.011
Amabile, A., Migliara, A., Capasso, P., Biffi, M., Cittaro, D., Naldini, L., et al. (2016). Inheritable silencing of endogenous genes by hit-and-run targeted epigenetic editing. Cell 167, 219–232.e14. doi: 10.1016/j.cell.2016.09.006
Argibay, B., Trekker, J., Himmelreich, U., Beiras, A., Topete, A., Taboada, P., et al. (2017). Intraarterial route increases the risk of cerebral lesions after mesenchymal cell administration in animal model of ischemia. Sci. Rep. 7:40758. doi: 10.1038/srep40758
Bailus, B. J., Pyles, B., McAlister, M. M., O’Geen, H., Lockwood, S. H., Adams, A. N., et al. (2016). Protein delivery of an artificial transcription factor restores widespread Ube3a expression in an Angelman syndrome mouse brain. Mol. Ther. 24, 548–555. doi: 10.1038/mt.2015.236
Barua, N. U., Gill, S. S., and Love, S. (2014). Convection-enhanced drug delivery to the brain: therapeutic potential and neuropathological considerations. Brain Pathol. 24, 117–127. doi: 10.1111/bpa.12082
Beegle, J., Hendrix, K., Maciel, H., Nolta, J. A., and Anderson, J. S. (2020). Improvement of motor and behavioral activity in Sandhoff mice transplanted with human CD34+ cells transduced with a HexA/HexB expressing Lentiviral vector. J. Gene Med. 22:e3205. doi: 10.1002/jgm.3205
Berg, E. L., Copping, N. A., Rivera, J. K., Pride, M. C., Careaga, M., Bauman, M. D., et al. (2018). Developmental social communication deficits in the Shank3 rat model of phelan-mcdermid syndrome and autism spectrum disorder. Autism. Res. 11, 587–601. doi: 10.1002/aur.1925
Berg, E. L., Pride, M. C., Petkova, S. P., Lee, R. D., Copping, N. A., Shen, Y., et al. (2020). Translational outcomes in a full gene deletion of ubiquitin protein ligase E3A rat model of Angelman syndrome. Transl. Psychiatry 10:39. doi: 10.1038/s41398-020-0720-2
Bolte, S., and Cordelières, F. P. (2006). A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213–232. doi: 10.1111/j.1365-2818.2006.01706.x
Born, H. A., Dao, A. T., Levine, A. T., Lee, W. L., Mehta, N. M., Mehra, S., et al. (2017). Strain-dependence of the Angelman syndrome phenotypes in Ube3a maternal deficiency mice. Sci. Rep. 7:8451. doi: 10.1038/s41598-017-08825-x
Brinker, T., Stopa, E., Morrison, J., and Klinge, P. (2014). A new look at cerebrospinal fluid circulation. Fluids Barr. CNS 11:10.
Chamberlain, S. J., and Lalande, M. (2010). Angelman syndrome, a genomic imprinting disorder of the brain. J. Neurosci. 30, 9958–9963. doi: 10.1523/JNEUROSCI.1728-10.2010
Chen, P.-J., Kang, Y.-D., Lin, C.-H., Chen, S.-Y., Hsieh, C.-H., Chen, Y.-Y., et al. (2015). Multitheragnostic Multi-GNRs crystal-seeded magnetic nanoseaurchin for enhanced in vivo mesenchymal-stem-cell homing, multimodal imaging, and stroke therapy. Adv. Mater. 27, 6488–6495. doi: 10.1002/adma.201502784
Cheng, Q., Wei, T., Farbiak, L., Johnson, L. T., Dilliard, S. A., and Siegwart, D. J. (2020). Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR–Cas gene editing. Nat. Nanotechnol. 15, 313–320. doi: 10.1038/s41565-020-0669-6
Damasceno, P. K. F., de Santana, T. A., Santos, G. C., Orge, I. D., Silva, D. N., Albuquerque, J. F., et al. (2020). Genetic engineering as a strategy to improve the therapeutic efficacy of mesenchymal Stem/Stromal cells in regenerative medicine. Front. Cell Dev. Biol. 8:737. doi: 10.3389/fcell.2020.00737
Dindot, S. V., Antalffy, B. A., Bhattacharjee, M. B., and Beaudet, A. L. (2008). The Angelman syndrome ubiquitin ligase localizes to the synapse and nucleus, and maternal deficiency results in abnormal dendritic spine morphology. Hum. Mol. Genet. 17, 111–118. doi: 10.1093/hmg/ddm288
Doeppner, T. R., El Aanbouri, M., Dietz, G. P. H., Weise, J., Schwarting, S., and Bähr, M. (2010). Transplantation of TAT-Bcl-xL-transduced neural precursor cells: long-term neuroprotection after stroke. Neurobiol. Dis. 40, 265–276. doi: 10.1016/j.nbd.2010.05.033
Fink, K. D., Deng, P., Gutierrez, J., Anderson, J. S., Torrest, A., Komarla, A., et al. (2016). Allele-specific reduction of the mutant huntingtin allele using transcription activator-like effectors in human huntington’s disease fibroblasts. Cell Transpl. 25, 677–686. doi: 10.3727/096368916X690863
Flinterman, M., Farzaneh, F., Habib, N., Malik, F., Gäken, J., and Tavassoli, M. (2009). Delivery of therapeutic proteins as secretable TAT fusion products. Mol. Ther. 17, 334–342.
Gilbert, L. A., Horlbeck, M. A., Adamson, B., Villalta, J. E., Chen, Y., Whitehead, E. H., et al. (2014). Genome-scale CRISPR-mediated control of gene repression and activation. Cell 159, 647–661. doi: 10.1016/j.cell.2014.09.029
Gong, B., Cao, Z., Zheng, P., Vitolo, O. V., Liu, S., Staniszewski, A., et al. (2006). Ubiquitin hydrolase Uch-L1 Rescues β-Amyloid-induced decreases in synaptic function and contextual memory. Cell 126, 775–788. doi: 10.1016/j.cell.2006.06.046
Halmai, J. A. N. M., Deng, P., Gonzalez, C. E., Coggins, N. B., Cameron, D., Carter, J. L., et al. (2020). Artificial escape from XCI by DNA methylation editing of the CDKL5 gene. Nucleic Acids Res. 48, 2372–2387. doi: 10.1093/nar/gkz1214
Hordeaux, J., Hinderer, C., Buza, E. L., Louboutin, J. P., Jahan, T., Bell, P., et al. (2019). Safe and sustained expression of human iduronidase after intrathecal administration of adeno-associated virus Serotype 9 in infant rhesus monkeys. Hum. Gene Ther. 30, 957–966. doi: 10.1089/hum.2019.012
Huang, H. S., Burns, A. J., Nonneman, R. J., Baker, L. K., Riddick, N. V., Nikolova, V. D., et al. (2013). Behavioral deficits in an Angelman syndrome model: effects of genetic background and age. Behav. Brain Res. 243, 79–90. doi: 10.1016/j.bbr.2012.12.052
Jiang, Y. H., Armstrong, D., Albrecht, U., Atkins, C. M., Noebels, J. L., Eichele, G., et al. (1998). Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron 21, 799–811. doi: 10.1016/s0896-6273(00)80596-6
Kalomoiris, S., Lawson, J., Chen, R. X., Bauer, G., Nolta, J. A., and Anderson, J. S. (2012). CD25 preselective Anti-HIV vectors for improved HIV Gene therapy. Hum. Gene Ther. Methods 23:366. doi: 10.1089/hgtb.2012.142
Kean, T. J., Lin, P., Caplan, A. I., and Dennis, J. E. (2013). MSCs: delivery routes and engraftment, cell-targeting strategies, and immune modulation. Stem Cells Intern. 2013:732742. doi: 10.1155/2013/732742
Keute, M., Miller, M. T., Krishnan, M. L., Sadhwani, A., Chamberlain, S., Thibert, R. L., et al. (2020). Angelman syndrome genotypes manifest varying degrees of clinical severity and developmental impairment. Mol. Psychiatry 26, 3625–3633. doi: 10.1038/s41380-020-0858-6
Kilic, E., Dietz, G. P. H., Hermann, D. M., and Bähr, M. (2002). Intravenous TAT-Bcl-XL is protective after middle cerebral artery occlusion in mice. Ann. Neurol. 52, 617–622. doi: 10.1002/ana.10356
Kilic, Ü, Kilic, E., Dietz, G. P. H., and Bähr, M. (2003). Intravenous TAT-GDNF is protective after focal cerebral ischemia in mice. Stroke 34, 1304–1310. doi: 10.1161/01.STR.0000066869.45310.50
Kishino, T., Lalande, M., and Wagstaff, J. (1997). UBE3A/E6-AP mutations cause Angelman syndrome. Nat. Genet. 15, 70–73. doi: 10.1038/ng0197-70
Kohn, D. B., Booth, C., Shaw, K. L., Xu-Bayford, J., Garabedian, E., Trevisan, V., et al. (2021). Autologous ex vivo Lentiviral gene therapy for adenosine deaminase deficiency. N. Engl. J. Med. 384, 2002–2013.
Kohn, D. B., Hershfield, M. S., Carbonaro, D., Shigeoka, A., Brooks, J., Smogorzewska, E. M., et al. (1998). Tlymphocytes with a normal ADA gene accumulate after transplantation of transduced autologous umbilical cord blood CD34+ cells in ADA-deficient SCID neonates. Nat. Med. 4, 775–780. doi: 10.1038/nm0798-775
Kohn, D. B., Weinberg, K. I., Nolta, J. A., Heiss, L. N., Lenarsky, C., Crooks, G. M., et al. (1995). Engraftment of gene–modified umbilical cord blood cells in neonates with adenosine deaminase deficiency. Nat. Med. 1, 1017–1023. doi: 10.1038/nm1095-1017
Ku, K. M., Weir, R. K., Silverman, J. L., Berman, R. F., and Bauman, M. D. (2016). Behavioral phenotyping of juvenile long-evans and sprague-dawley rats: implications for preclinical models of autism spectrum disorders. PLoS One 11:e0158150. doi: 10.1371/journal.pone.0158150
Landers, M., Bancescu, D. L., Le Meur, E., Rougeulle, C., Glatt-Deeley, H., Brannan, C., et al. (2004). Regulation of the large (∼1000 kb) imprinted murine Ube3a antisense transcript by alternative exons upstream of Snurf/Snrpn. Nucleic Acids Res. 32, 3480–3492. doi: 10.1093/nar/gkh670
Lee, C. C. I., Ye, F., and Tarantal, A. F. (2006). Comparison of growth and differentiation of fetal and adult rhesus monkey mesenchymal stem cells. Stem Cells Dev. 15, 209–220. doi: 10.1089/scd.2006.15.209
Lee, C. I., Kohn, D. B., Ekert, J. E., and Tarantal, A. F. (2004). Morphological analysis and Lentiviral transduction of fetal monkey bone marrow-derived mesenchymal stem cells. Mol. Ther. 9, 112–123. doi: 10.1016/j.ymthe.2003.09.019
Liang, X., Ding, Y., Zhang, Y., Tse, H. F., and Lian, Q. (2014). Paracrine mechanisms of mesenchymal stem cell-based therapy: current status and perspectives. Cell Transpl. 23, 1045–1059. doi: 10.3727/096368913X667709
Lu, M., Chen, J., Lu, D., Yi, L., Mahmood, A., and Chopp, M. (2003). Global test statistics for treatment effect of stroke and traumatic brain injury in rats with administration of bone marrow stromal cells. J. Neurosci. Methods 128, 183–190. doi: 10.1016/s0165-0270(03)00188-2
Mazur, C., Powers, B., Zasadny, K., Sullivan, J. M., Dimant, H., Kamme, F., et al. (2019). Brain pharmacology of intrathecal antisense oligonucleotides revealed through multimodal imaging. JCI Insight. 4:e129240. doi: 10.1172/jci.insight.129240
Mendell, J. R., Al-Zaidy, S., Shell, R., Arnold, W. D., Rodino-Klapac, L. R., Prior, T. W., et al. (2017). Single-dose gene-replacement therapy for spinal muscular atrophy. N. Engl. J. Med. 377, 1713–1722. doi: 10.1056/NEJMoa1706198
Meyerrose, T., Olson, S., Pontow, S., Kalomoiris, S., Jung, Y., Annett, G., et al. (2010). Mesenchymal stem cells for the sustained in vivo delivery of bioactive factors. Adv. Drug Deliv. Rev. 62, 1167–1174. doi: 10.1016/j.addr.2010.09.013
Nuñez, J. K., Chen, J., Pommier, G. C., Cogan, J. Z., Replogle, J. M., Adriaens, C., et al. (2021). Genome-wide programmable transcriptional memory by CRISPR-based epigenome editing. Cell 184, 2503–2519.e17. doi: 10.1016/j.cell.2021.03.025
O’Geen, H., Bates, S. L., Carter, S. S., Nisson, K. A., Halmai, J., Fink, K. D., et al. (2019). Ezh2-dCas9 and KRAB-dCas9 enable engineering of epigenetic memory in a context-dependent manner. Epigenet. Chrom. 12:26. doi: 10.1186/s13072-019-0275-8
Olabarria, M., Pasini, S., Corona, C., Robador, P., Song, C., Patel, H., et al. (2019). Dysfunction of the ubiquitin ligase E3A Ube3A/E6-AP contributes to synaptic pathology in Alzheimer’s disease. Commun. Biol. 2:111. doi: 10.1038/s42003-019-0350-5
Olson, S. D., Kambal, A., Pollock, K., Mitchell, G. M., Stewart, H., Kalomoiris, S., et al. (2012). Examination of mesenchymal stem cell-mediated RNAi transfer to Huntington’s disease affected neuronal cells for reduction of huntingtin. Mol. Cell Neurosci. 49, 271–281. doi: 10.1016/j.mcn.2011.12.001
Papisov, M. I., Belov, V., Fischman, A. J., Belova, E., Titus, J., Gagne, M., et al. (2012). Delivery of proteins to CNS as seen and measured by positron emission tomography. Drug Deliv. Transl. Res. 2, 201–209. doi: 10.1007/s13346-012-0073-3
Perdigão, P. R. L., Cunha-Santos, C., Barbas, C. F., Santa-Marta, M., and Goncalves, J. (2020). Protein delivery of cell-penetrating zinc-finger activators stimulates latent HIV-1-infected cells. Mol. Ther. Methods Clin. Dev. 18, 145–158. doi: 10.1016/j.omtm.2020.05.016
Pollock, K., Dahlenburg, H., Nelson, H., Fink, K. D., Cary, W., Hendrix, K., et al. (2016). Human mesenchymal stem cells genetically engineered to overexpress brain-derived neurotrophic factor improve outcomes in huntington’s disease mouse models. Mol. Ther. 24, 965–977. doi: 10.1038/mt.2016.12
Ramakrishna, S., Kim, Y. H., and Kim, H. (2013). Stability of zinc finger nuclease protein is enhanced by the proteasome inhibitor MG132. PLoS One 8:e54282. doi: 10.1371/journal.pone.0054282
Rossignol, J., Boyer, C., Lévèque, X., Fink, K. D., Thinard, R., Blanchard, F., et al. (2011). Mesenchymal stem cell transplantation and DMEM administration in a 3NP rat model of Huntington’s disease: Morphological and behavioral outcomes. Behav. Brain Res. 217, 369–378. doi: 10.1016/j.bbr.2010.11.006
Russ, A. P., and Lampel, S. (2005). The druggable genome: an update. Drug Discov. Today 10, 1607–1610. doi: 10.1016/S1359-6446(05)03666-4
Russell, S., Bennett, J., Wellman, J. A., Chung, D. C., Yu, Z. F., Tillman, A., et al. (2017). Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: a randomised, controlled, open-label, phase 3 trial. Lancet 390, 849–860. doi: 10.1016/S0140-6736(17)31868-8
Schwarze, S. R., Ho, A., Vocero-Akbani, A., and Dowdy, S. F. (1999). In vivo protein transduction: delivery of a biologically active protein into the mouse. Science 285, 1569–1572. doi: 10.1126/science.285.5433.1569
Silva-Santos, S., Van Woerden, G. M., Bruinsma, C. F., Mientjes, E., Jolfaei, M. A., Distel, B., et al. (2015). Ube3a reinstatement identifies distinct developmental windows in a murine Angelman syndrome model. J. Clin. Invest. 125, 2069–2076. doi: 10.1172/JCI80554
Sonzogni, M., Wallaard, I., Santos, S. S., Kingma, J., Du Mee, D., Van Woerden, G. M., et al. (2018). A behavioral test battery for mouse models of Angelman syndrome: a powerful tool for testing drugs and novel Ube3a mutants. Mol. Autism. 9:47. doi: 10.1186/s13229-018-0231-7
Tan, W.-H., Bacino, C. A., Skinner, S. A., Anselm, I., Barbieri-Welge, R., Bauer-Carlin, A., et al. (2011). Angelman syndrome: mutations influence features in early childhood. Am. J. Med. Genet. Part A 155, 81–90. doi: 10.1002/ajmg.a.33775
Tarantal, A. F., and Lee, C. C. I. (2010). Long-term luciferase expression monitored by bioluminescence imaging after adeno-associated virus-mediated fetal gene delivery in rhesus monkeys (Macaca mulatta). Hum. Gene Ther. 21, 143–148. doi: 10.1089/hum.2009.126
Tarantal, A. F., Lee, C. C. I., and Itkin-Ansari, P. (2009). Real-time bioluminescence imaging of macroencapsulated fibroblasts reveals allograft protection in rhesus monkeys (Macaca mulatta). Transplantation 88, 38–41. doi: 10.1097/TP.0b013e3181a9ee6c
Thakore, P. I., D’Ippolito, A. M., Song, L., Safi, A., Shivakumar, N. K., Kabadi, A. M., et al. (2015). Highly specific epigenome editing by CRISPR-Cas9 repressors for silencing of distal regulatory elements. Nat. Methods 12, 1143–1149. doi: 10.1038/nmeth.3630
Thompson, M., Mei, S. H. J., Wolfe, D., Champagne, J., Fergusson, D., Stewart, D. J., et al. (2020). Cell therapy with intravascular administration of mesenchymal stromal cells continues to appear safe: an updated systematic review and meta-analysis. EClin. Med. 19:100249. doi: 10.1016/j.eclinm.2019.100249
Visigalli, I., Delai, S., Politi, L. S., Di Domenico, C., Cerri, F., Mrak, E., et al. (2010). Gene therapy augments the efficacy of hematopoietic cell transplantation and fully corrects mucopolysaccharidosis type I phenotype in the mouse model. Blood 116, 5130–5139. doi: 10.1182/blood-2010-04-278234
Wang, M., Zuris, J. A., Meng, F., Rees, H., Sun, S., Deng, P., et al. (2016). Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles. Proc. Natl. Acad. Sci. U.S.A. 113, 2868–2873. doi: 10.1073/pnas.1520244113
Wheeler, A. C., Sacco, P., and Cabo, R. (2017). Unmet clinical needs and burden in Angelman syndrome: a review of the literature. Orphanet. J. Rare Dis. 12:164. doi: 10.1186/s13023-017-0716-z
Williams, C. A. (2010). The behavioral phenotype of the Angelman syndrome. Am. J. Med. Genet. Part C Semin. Med. Genet. 154, 432–437. doi: 10.1002/ajmg.c.30278
Wolter, J. M., Mao, H., Fragola, G., Simon, J. M., Krantz, J. L., Bazick, H. O., et al. (2020). Cas9 gene therapy for Angelman syndrome traps Ube3a-ATS long non-coding RNA. Nature 587, 281–284. doi: 10.1038/s41586-020-2835-2
Wu, Y., Luo, X., Liu, X., Liu, D., Wang, X., Guo, Z., et al. (2015). Intraperitoneal administration of a novel TAT-BDNF peptide ameliorates cognitive impairments via modulating multiple pathways in two Alzheimer’s rodent models. Sci. Rep. 5:15032. doi: 10.1038/srep15032
Keywords: zinc finger, Angelman Syndrome (AS), mesenchymal stem/stromal cell, artificial transcription factor (ATF), cell-based delivery
Citation: Deng P, Halmai JANM, Beitnere U, Cameron D, Martinez ML, Lee CC, Waldo JJ, Thongphanh K, Adhikari A, Copping N, Petkova SP, Lee RD, Lock S, Palomares M, O’Geen H, Carter J, Gonzalez CE, Buchanan FKB, Anderson JD, Fierro FA, Nolta JA, Tarantal AF, Silverman JL, Segal DJ and Fink KD (2022) An in vivo Cell-Based Delivery Platform for Zinc Finger Artificial Transcription Factors in Pre-clinical Animal Models. Front. Mol. Neurosci. 14:789913. doi: 10.3389/fnmol.2021.789913
Received: 05 October 2021; Accepted: 01 December 2021;
Published: 27 January 2022.
Edited by:
Johannes Boltze, University of Warwick, United KingdomReviewed by:
Xiao-Hong Lu, Louisiana State University Health Shreveport, United StatesSudhanshu P. Raikwar, Barrow Neurological Institute (BNI), United States
Copyright © 2022 Deng, Halmai, Beitnere, Cameron, Martinez, Lee, Waldo, Thongphanh, Adhikari, Copping, Petkova, Lee, Lock, Palomares, O’Geen, Carter, Gonzalez, Buchanan, Anderson, Fierro, Nolta, Tarantal, Silverman, Segal and Fink. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kyle D. Fink, kdfink@ucdavis.edu
†Lead Contact
 Peter Deng
Peter Deng Julian A. N. M. Halmai
Julian A. N. M. Halmai Ulrika Beitnere
Ulrika Beitnere David Cameron1,2,4
David Cameron1,2,4  Krista Thongphanh
Krista Thongphanh Anna Adhikari
Anna Adhikari Samantha Lock
Samantha Lock Henriette O’Geen
Henriette O’Geen Jasmine Carter
Jasmine Carter Johnathan D. Anderson
Johnathan D. Anderson Alice F. Tarantal
Alice F. Tarantal Jill L. Silverman
Jill L. Silverman David J. Segal
David J. Segal Kyle D. Fink
Kyle D. Fink